More Manipulations of Land Cover Raster
Elie and Laura
2019-09-27
This is a primer for manipulating a Canada-wide land cover raster of the Northwest Territories. This raster is not as detailed as the EOSD, but it provides an opportuntity to practice manipulating rasters. In particular, we DEFINE and COLLAPSE categorical layers.
1 Load the Land Cover
The land cover raster we are using is from the North American Land Cover Management System (NALCMS), a joint project between Canadian, US, and Mexican organizations. The project is a multi-scale land cover monitoring framework which can be applied across North America. This raster is of a small section of the Northwest Territories, Canada, on the western edge of Great Slave Lake.
Load the raster with the raster command:
require(raster)
landcover <- raster("_data/LandCover/LandCovCanada_c2010_NALCMSlegend_HRL_clip.tif")
landcover## class : RasterLayer
## dimensions : 10290, 10687, 109969230 (nrow, ncol, ncell)
## resolution : 30, 30 (x, y)
## extent : -1358210, -1037600, 7962570, 8271270 (xmin, xmax, ymin, ymax)
## crs : +proj=lcc +lat_1=49 +lat_2=77 +lat_0=0 +lon_0=-95 +x_0=0 +y_0=0 +ellps=GRS80 +units=m +no_defs
## source : C:/Users/Elie/Box Sync/caribou/NWT/rsf/primers/_data/LandCover/LandCovCanada_c2010_NALCMSlegend_HRL_clip.tif
## names : LandCovCanada_c2010_NALCMSlegend_HRL_clip
## values : 1, 18 (min, max)
## attributes :
## ID OBJECTID Value Count
## from: 0 1 1 24924814
## to : 11 12 18 4645750It plots nicely:
plot(landcover)
But there are no classifications: the raster has an attribute table, but the names of the land cover types are not specified. We need to match up the “Values” to their respective land cover types.
levels(landcover)## [[1]]
## ID OBJECTID Value Count
## 1 0 1 1 24924814
## 2 1 2 2 1973279
## 3 2 3 5 1192339
## 4 3 4 6 2274680
## 5 4 5 8 4593568
## 6 5 6 10 55546
## 7 6 7 11 74603
## 8 7 8 12 692538
## 9 8 9 14 4990755
## 10 9 10 16 7184
## 11 10 11 17 31523
## 12 11 12 18 4645750These same land cover categories are used throughout the NALCMS dataset, so we can pull the legend from their website: https://landcover.usgs.gov/nalcms.php Unfortunately the legend is in .jpg format, but it’s easy enough to copy into an excel file, which we did manually.
| Value | Description | R | G | B |
|---|---|---|---|---|
| 1 | Temperate or sub-polar needleleaf forest | 0 | 61 | 1 |
| 2 | Sub-polar taiga needleleaf forest | 147 | 156 | 113 |
| 3 | Tropical or sub-tropical broadleaf evergreen forest | 1 | 99 | 0 |
| 4 | Tropical or sub-tropical broadleaf deciduous forest | 29 | 170 | 4 |
| 5 | Temperate or sub-polar broadleaf deciduous forest | 19 | 140 | 61 |
| 6 | Mixed Forest | 91 | 117 | 43 |
| 7 | Tropical or sub-tropical Shrubland | 178 | 158 | 43 |
| 8 | Temperate or sub-polar Shrubland | 177 | 137 | 50 |
| 9 | Tropical or sub-tropical Grassland | 233 | 219 | 94 |
| 10 | Temperate or sub-polar Grassland | 225 | 206 | 137 |
| 11 | Sub-polar or polar shrubland-lichen-moss | 156 | 117 | 84 |
| 12 | Sub-polar or polar grassland-lichen-moss | 186 | 211 | 143 |
| 13 | Sub-polar or polar barren-lichen-moss | 63 | 137 | 112 |
| 14 | Wetland | 107 | 163 | 136 |
| 15 | Cropland | 230 | 173 | 102 |
| 16 | Barren land | 169 | 170 | 174 |
| 17 | Urban | 219 | 33 | 38 |
| 18 | Water | 76 | 112 | 164 |
| 19 | Snow and Ice | 255 | 248 | 255 |
Here is a useful website that can be used to find the specific RGB, HEX or CYMK code of any color in an image. I used it to pull out the RGB code for all the colors in the legend .jpg file: https://www.ginifab.com/feeds/pms/pms_color_in_image.php
2 Add a legend to the Raster:
In order to attach the legend to our raster, we need to merge the levels of the landcover raster with the legend dataframe. Both files have a column named “Value” which we will base the merge on:
levels(landcover)[[1]] <- merge(levels(landcover)[[1]], legend, by = "Value")[,c(2,1,3:5)]Now we add the colors. The correct colortable is already present in the landcover raster. Here we take the colors in the colortable and add them as a new column in the raster attribute table.(See examples from primer 1)
Note: the raster’s colortable is offset by 1 in relation to our imported legend (the color of the first landcover type is actually the second entry in the list). To keep everything lined up correctly we use
Value + 1instead of just the value.
levels(landcover)[[1]]$Colors <- landcover@legend@colortable[levels(landcover)[[1]]$Value + 1]And there you have it:
plot(landcover)
legend("topright", fill = levels(landcover)[[1]]$Color, legend = levels(landcover)[[1]]$Description, ncol = 1, bg = "white", cex = 0.6)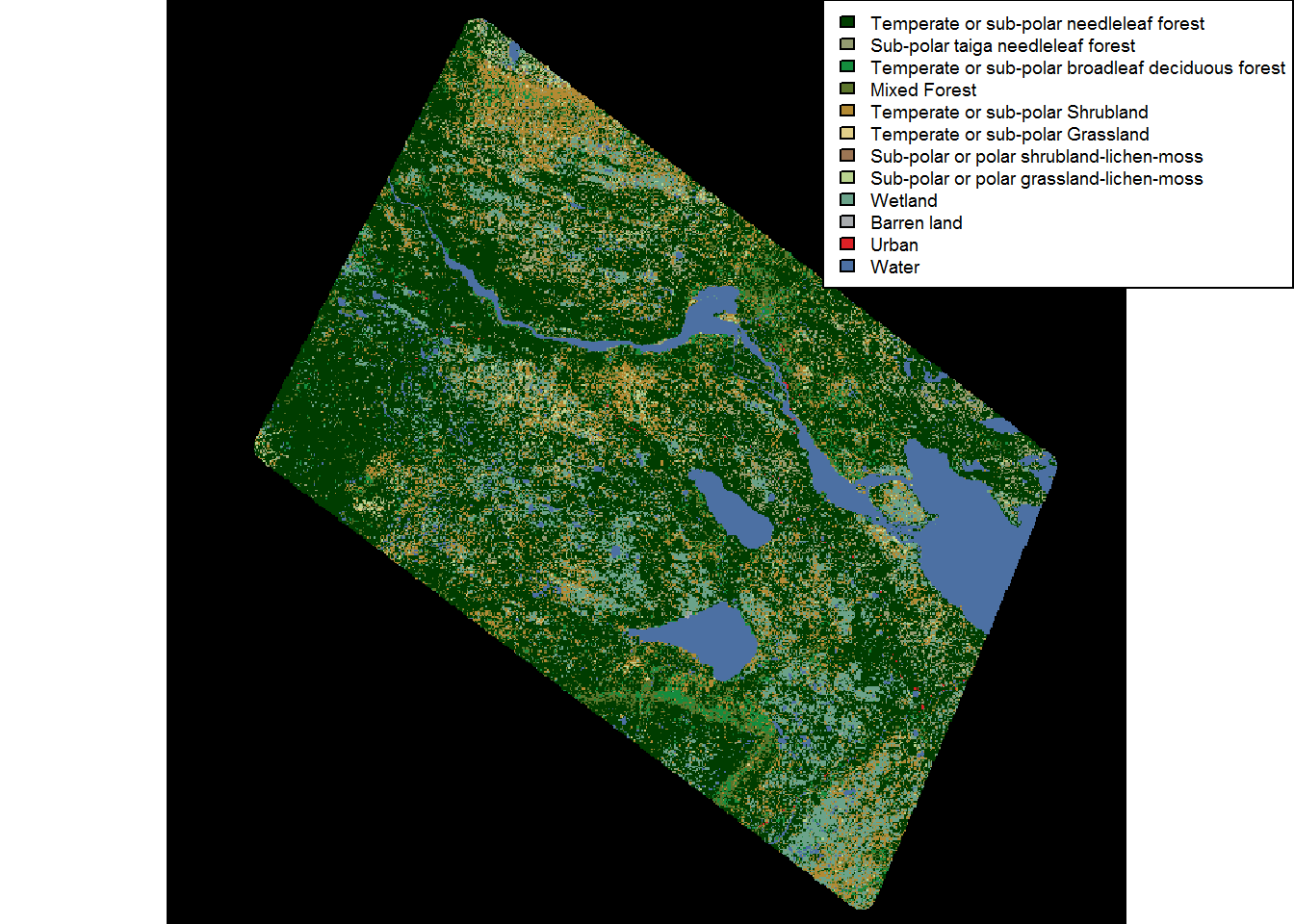
3 Pool classes together
We have a lot of different land cover types here. To simplify things we can combine similar types into one large land cover category. For example, let’s pool both Temperate or sub-polar needleleaf forest and Sub-polar taiga needleleaf forest (values 17 and 18) into a single class called “1: Needleleaf Forest”.
Steps are simple:
- Turn all 2’s into 1’s
- Rename 1
- Remove row for 2
landcover.pooled <- landcover
values(landcover.pooled)[values(landcover.pooled) == 2] <- 1
mylevels <- levels(landcover.pooled)[[1]]
mylevels$Description <- as.character(mylevels$Description)
mylevels[mylevels$Value == 1,]$Description <- "Needleleaf Forest"
mylevels <- subset(mylevels, Value != 2)
levels(landcover.pooled)[[1]] <- mylevelsTo make this more general, e.g. to pool values from a vector sourcevalues to a new value targetvalue and new description newdescription, we write a function:
poolValues <- function(raster, sourcevalues, targetvalue, newdescription){
raster.pooled <- raster
values(raster.pooled)[values(raster.pooled) %in% sourcevalues] <- targetvalue
mylevels <- levels(raster.pooled)[[1]]
mylevels$Description = as.character(mylevels$Description)
mylevels[mylevels$Value == targetvalue,]$Description <- newdescription
mylevels <- subset(mylevels, !(Value %in% sourcevalues) | Value == targetvalue )
levels(raster.pooled)[[1]] <- suppressWarnings(mylevels)
return(raster.pooled)
}Note that this function works on any raster, as long as there is a Description column in the levels.
Let’s test this by pooling all of the Forests (classes 1, 2, 5 and 6) into a single monolithic FOREST class:
landcover.forestpooled <- poolValues(landcover, c(1,2,5,6), 1, "Forest")
plot(landcover.forestpooled)
legend("topright", fill = levels(landcover.forestpooled)[[1]]$Color,
legend = levels(landcover.forestpooled)[[1]]$Description,
ncol = 1, bg = "white", cex = 0.6)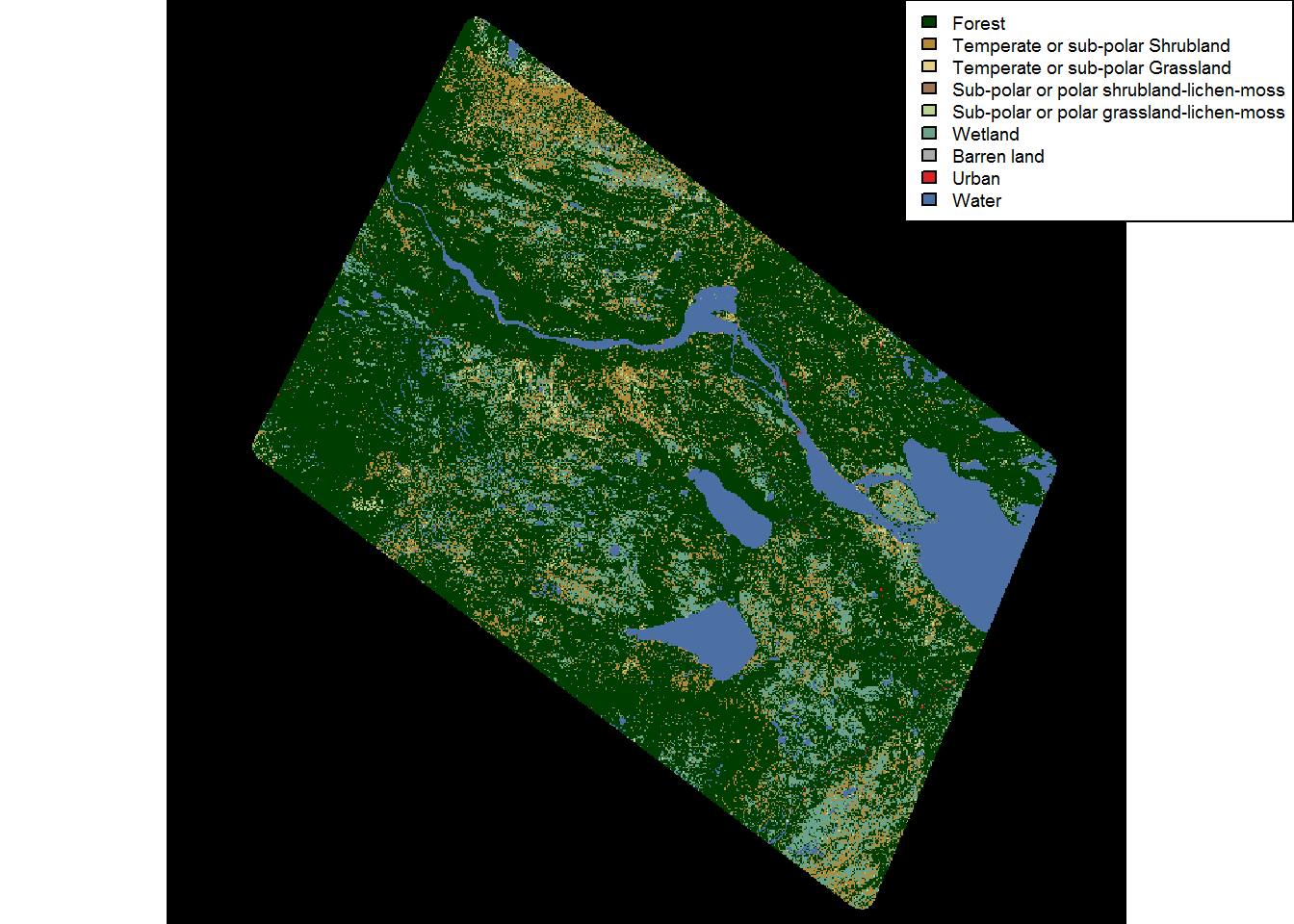
A lot of the forest nuance is now gone! For caribou - probably too much.
| ID | Value | OBJECTID | Count | Description | Colors | |
|---|---|---|---|---|---|---|
| 1 | 0 | 1 | 1 | 24924814 | Forest | #003D00 |
| 5 | 4 | 8 | 5 | 4593568 | Temperate or sub-polar Shrubland | #B38A33 |
| 6 | 5 | 10 | 6 | 55546 | Temperate or sub-polar Grassland | #E1CF8A |
| 7 | 6 | 11 | 7 | 74603 | Sub-polar or polar shrubland-lichen-moss | #9C7554 |
| 8 | 7 | 12 | 8 | 692538 | Sub-polar or polar grassland-lichen-moss | #BAD48F |
| 9 | 8 | 14 | 9 | 4990755 | Wetland | #6BA38A |
| 10 | 9 | 16 | 10 | 7184 | Barren land | #A8ABAE |
| 11 | 10 | 17 | 11 | 31523 | Urban | #DC2126 |
| 12 | 11 | 18 | 12 | 4645750 | Water | #4C70A3 |
Note: Although there is a column in the dataframe named “Count” this doesn’t actually COUNT anything! Now that multiple rows have been pooled together the count listed in the dataframe isn’t necesarily correct.If you want to know the count (the number of pixels in each land cover category) that would have to be done with the
tablefunction (which - for a large raster - can be pretty slow). Below, the count is correct:
levels(landcover.forestpooled)[[1]]$Count <- table(getValues(landcover.forestpooled))| ID | Value | OBJECTID | Count | Description | Colors | |
|---|---|---|---|---|---|---|
| 1 | 0 | 1 | 1 | 30365112 | Forest | #003D00 |
| 5 | 4 | 8 | 5 | 4593568 | Temperate or sub-polar Shrubland | #B38A33 |
| 6 | 5 | 10 | 6 | 55546 | Temperate or sub-polar Grassland | #E1CF8A |
| 7 | 6 | 11 | 7 | 74603 | Sub-polar or polar shrubland-lichen-moss | #9C7554 |
| 8 | 7 | 12 | 8 | 692538 | Sub-polar or polar grassland-lichen-moss | #BAD48F |
| 9 | 8 | 14 | 9 | 4990755 | Wetland | #6BA38A |
| 10 | 9 | 16 | 10 | 7184 | Barren land | #A8ABAE |
| 11 | 10 | 17 | 11 | 31523 | Urban | #DC2126 |
| 12 | 11 | 18 | 12 | 4645750 | Water | #4C70A3 |
4 Complete code
Click below to show complete code from this document.
knitr::opts_chunk$set(echo = TRUE, cache=TRUE, message = FALSE, warning = FALSE)
require(raster)
landcover <- raster("_data/LandCover/LandCovCanada_c2010_NALCMSlegend_HRL_clip.tif")
landcover
plot(landcover)
levels(landcover)
require(knitr)
legend <- read.csv("_data/LandCover/NALCMS_Legend.csv")
names(legend)[1] <- "Value"
kable(legend)
levels(landcover)[[1]] <- merge(levels(landcover)[[1]], legend, by = "Value")[,c(2,1,3:5)]
levels(landcover)[[1]]$Colors <- landcover@legend@colortable[levels(landcover)[[1]]$Value + 1]
plot(landcover)
legend("topright", fill = levels(landcover)[[1]]$Color, legend = levels(landcover)[[1]]$Description, ncol = 1, bg = "white", cex = 0.6)
landcover.pooled <- landcover
values(landcover.pooled)[values(landcover.pooled) == 2] <- 1
mylevels <- levels(landcover.pooled)[[1]]
mylevels$Description <- as.character(mylevels$Description)
mylevels[mylevels$Value == 1,]$Description <- "Needleleaf Forest"
mylevels <- subset(mylevels, Value != 2)
levels(landcover.pooled)[[1]] <- mylevels
poolValues <- function(raster, sourcevalues, targetvalue, newdescription){
raster.pooled <- raster
values(raster.pooled)[values(raster.pooled) %in% sourcevalues] <- targetvalue
mylevels <- levels(raster.pooled)[[1]]
mylevels$Description = as.character(mylevels$Description)
mylevels[mylevels$Value == targetvalue,]$Description <- newdescription
mylevels <- subset(mylevels, !(Value %in% sourcevalues) | Value == targetvalue )
levels(raster.pooled)[[1]] <- suppressWarnings(mylevels)
return(raster.pooled)
}
landcover.forestpooled <- poolValues(landcover, c(1,2,5,6), 1, "Forest")
plot(landcover.forestpooled)
legend("topright", fill = levels(landcover.forestpooled)[[1]]$Color,
legend = levels(landcover.forestpooled)[[1]]$Description,
ncol = 1, bg = "white", cex = 0.6)
require(knitr); require(magrittr)
levels(landcover.forestpooled)[[1]] %>% kable
levels(landcover.forestpooled)[[1]]$Count <- table(getValues(landcover.forestpooled))
levels(landcover.forestpooled)[[1]] %>% kable