Step 01: Loading and Processing Movebank Data
Elie Gurarie
July 27, 2018
Goal: to load a movement study from Movebank, obtain the ancillary individual ID data, download additional individual specific data, and link to the movebank data. We also cover some basic plotting and mapping of the movebank data. We will work with data from the Inuvik woodland population (follow link to open the study page on Movebank).
1 Loading data from Movebank
From within R, load the move package:
require(move)Create a login object, using your own movebank.org login and password, as follows:
mylogin <- movebankLogin(username="MyUserName", password="MyPassword")To load a study, it is important that it be spelled out correctly, exactly as it is called in the movebank page. The following command will load the Inuvik data:
inuvik <- getMovebankData("ABoVE: NWT Inuvik Boreal Woodland Caribou", login = mylogin)## Error in curl::curl_fetch_memory(url, handle = handle): Could not resolve host: www.movebank.orgThis step might take some time. But when it is done you will have a new object, called inuvik in yuor workspace. This is the entirety of that study’s data. You will want to save this object locally, as an R object, so you can load it immediately as needed:
save(inuvik, file = "./_data/inuvik.rda")2 Structure of movebank data
The structure of the inuvik object is rather complex. Entering inuvik into the console will spit out a lot of information:
inuvik## class : MoveStack
## features : 69997
## extent : -136.024, -129.7156, 66.134, 68.8489 (xmin, xmax, ymin, ymax)
## coord. ref. : +proj=longlat +ellps=WGS84 +datum=WGS84 +towgs84=0,0,0
## variables : 15
## names : sensor_type_id, comments, habitat, import_marked_outlier, location_lat, location_long, manually_marked_outlier, timestamp, update_ts, argos_calcul_freq, argos_lc, deployment_id, event_id, tag_id, sensor_type
## min values : 653, NA, NA, NA, 66.1340, -129.7156, false, 2002-05-02 00:00:00, 2018-09-17 20:32:20.291, 401648030, 1, 532845467, 7411868493, 532618681, Argos Doppler Shift
## max values : 82798, NA, NA, NA, 68.8489, -136.0240, false, 2012-07-05 16:01:55, 2018-09-17 20:35:37.738, 401652084, 3, 532845511, 7411957236, 532839235, GPS
## timestamps : 2002-05-02 00:00:00 ... 2012-07-05 16:01:55 Time difference of 3718 days (start ... end, duration)
## sensors : GPS, Argos Doppler Shift
## indiv. data : comments, death_comments, earliest_date_born, exact_date_of_birth, individual_id, latest_date_born, local_identifier, nick_name, ring_id, sex, taxon_canonical_name
## min ID Data : NA, NA, NA, NA, 532618680, NA, BW26, IN0701, NA, f, caribou
## max ID Data : NA, NA, NA, NA, 532622218, NA, BWC36189, IN45, NA, f, caribou
## individuals : BW26, BW27, BW28, BW29, BW30, BW31, BW32, BW33, BW34, BW35, BW36, BW37, BW39, BW40, BW41
## unused rec. : 243
## license : Data are for use only as part of NASA's Arctic-Boreal Vul ...
## citation : Nagy JA, Derocher AE, Nielsen SE, Wright WH, Heikkila JM (2006) Modelling seasonal habitats of boreal woodland caribou at the northern limits of their range: a preliminary assessment of the Lower Mackenzie River Valley, Northwest Territories, Canada. Government of the Northwest Territories. <br><br> Nagy JA, Auriat D, Wright W, Slack T, Ellsworth I, Kienzler M (2004) Ecology of boreal woodland caribou in the Lower Mackenzie Valley NT: work completed in the Inuvik region April 2003 to November 2004. <br><br> See others at www.enr.gov.nt.ca/resources.
## study name : ABoVE: NWT Inuvik Boreal Woodland Caribou
## date created: 2018-09-14 21:44:18In fact, this is a movestack object, which contains information inside of elements called slots. Eg: a dataframe has column, but a movestack is an S4 “formal” dataclass which has strictly defined slots. This data structure is typical for spatial data.
2.1 Slots
To see the names of the slots:
slotNames(inuvik)## [1] "trackId" "timestamps"
## [3] "idData" "sensor"
## [5] "data" "coords.nrs"
## [7] "coords" "bbox"
## [9] "proj4string" "trackIdUnUsedRecords"
## [11] "timestampsUnUsedRecords" "sensorUnUsedRecords"
## [13] "dataUnUsedRecords" "dateCreation"
## [15] "study" "citation"
## [17] "license"You can access slots with the @ tag (contrast to the $ for data frames and lists). The data slot contains the movement data:
head(inuvik@data)| sensor_type_id | comments | habitat | import_marked_outlier | location_lat | location_long | manually_marked_outlier | timestamp | update_ts | argos_calcul_freq | argos_lc | deployment_id | event_id | tag_id | sensor_type | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 25915 | 653 | NA | NA | NA | 67.5443 | -132.1738 | false | 2005-03-11 00:00:00 | 2018-09-17 20:35:37.738 | NA | NA | 532845469 | 7411907216 | 532618725 | GPS |
| 25916 | 653 | NA | NA | NA | 67.5447 | -132.1735 | false | 2005-03-11 08:03:42 | 2018-09-17 20:35:37.738 | NA | NA | 532845469 | 7411907218 | 532618725 | GPS |
| 25917 | 653 | NA | NA | NA | 67.5448 | -132.1736 | false | 2005-03-11 16:01:55 | 2018-09-17 20:35:37.738 | NA | NA | 532845469 | 7411907217 | 532618725 | GPS |
| 25918 | 653 | NA | NA | NA | 67.5434 | -132.1618 | false | 2005-03-12 00:00:00 | 2018-09-17 20:35:37.738 | NA | NA | 532845469 | 7411907219 | 532618725 | GPS |
| 25919 | 653 | NA | NA | NA | 67.5432 | -132.1503 | false | 2005-03-12 07:58:05 | 2018-09-17 20:35:37.738 | NA | NA | 532845469 | 7411907221 | 532618725 | GPS |
| 25920 | 653 | NA | NA | NA | 67.5432 | -132.1504 | false | 2005-03-12 16:01:55 | 2018-09-17 20:35:37.738 | NA | NA | 532845469 | 7411907220 | 532618725 | GPS |
Other slots contain, e.g., the projection, the license, a citation:
inuvik@proj4string## CRS arguments:
## +proj=longlat +ellps=WGS84 +datum=WGS84 +towgs84=0,0,0inuvik@license## [1] "Data are for use only as part of NASA's Arctic-Boreal Vulnerability Experiment (ABoVE, limited to select ABoVE researchers) and by NWT staff."inuvik@citation## [1] "Nagy JA, Derocher AE, Nielsen SE, Wright WH, Heikkila JM (2006) Modelling seasonal habitats of boreal woodland caribou at the northern limits of their range: a preliminary assessment of the Lower Mackenzie River Valley, Northwest Territories, Canada. Government of the Northwest Territories. <br><br> Nagy JA, Auriat D, Wright W, Slack T, Ellsworth I, Kienzler M (2004) Ecology of boreal woodland caribou in the Lower Mackenzie Valley NT: work completed in the Inuvik region April 2003 to November 2004. <br><br> See others at www.enr.gov.nt.ca/resources."2.2 Individual ID information
The individual ID data is found in a “slot” called idData, with one row per individual:
inuvik@idData| comments | death_comments | earliest_date_born | exact_date_of_birth | individual_id | latest_date_born | local_identifier | nick_name | ring_id | sex | taxon_canonical_name | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| BW26 | NA | NA | NA | NA | 532618724 | NA | BW26 | IN26 | NA | f | caribou |
| BW27 | NA | NA | NA | NA | 532618742 | NA | BW27 | IN27 | NA | f | caribou |
| BW28 | NA | NA | NA | NA | 532618733 | NA | BW28 | IN28 | NA | f | caribou |
| BW29 | NA | NA | NA | NA | 532618736 | NA | BW29 | IN29 | NA | f | caribou |
| BW30 | NA | NA | NA | NA | 532618739 | NA | BW30 | IN30 | NA | f | caribou |
| BW31 | NA | NA | NA | NA | 532618729 | NA | BW31 | IN31 | NA | f | caribou |
| BW32 | NA | NA | NA | NA | 532622170 | NA | BW32 | IN32 | NA | f | caribou |
| BW33 | NA | NA | NA | NA | 532622185 | NA | BW33 | IN33 | NA | f | caribou |
| BW34 | NA | NA | NA | NA | 532618746 | NA | BW34 | IN34 | NA | f | caribou |
| BW35 | NA | NA | NA | NA | 532622188 | NA | BW35 | IN35 | NA | f | caribou |
| BW36 | NA | NA | NA | NA | 532622191 | NA | BW36 | IN36 | NA | f | caribou |
| BW37 | NA | NA | NA | NA | 532622194 | NA | BW37 | IN37 | NA | f | caribou |
| BW39 | NA | NA | NA | NA | 532622197 | NA | BW39 | IN39 | NA | f | caribou |
| BW40 | NA | NA | NA | NA | 532622200 | NA | BW40 | IN40 | NA | f | caribou |
| BW41 | NA | NA | NA | NA | 532622203 | NA | BW41 | IN41 | NA | f | caribou |
| BW42 | NA | NA | NA | NA | 532622206 | NA | BW42 | IN42 | NA | f | caribou |
| BW43 | NA | NA | NA | NA | 532618749 | NA | BW43 | IN43 | NA | f | caribou |
| BW44 | NA | NA | NA | NA | 532618799 | NA | BW44 | IN44 | NA | f | caribou |
| BW45 | NA | NA | NA | NA | 532618765 | NA | BW45 | IN45 | NA | f | caribou |
| BWC07.01 | NA | NA | NA | NA | 532618770 | NA | BWC07/01 | IN0701 | NA | f | caribou |
| BWC07.04 | NA | NA | NA | NA | 532618773 | NA | BWC07/04 | IN0704 | NA | f | caribou |
| BWC07.05 | NA | NA | NA | NA | 532618784 | NA | BWC07/05 | IN0705 | NA | f | caribou |
| BWC07.09 | NA | NA | NA | NA | 532618790 | NA | BWC07/09 | IN0709 | NA | f | caribou |
| BWC08.01 | NA | NA | NA | NA | 532618680 | NA | BWC08/01 | IN0801 | NA | f | caribou |
| BWC08.02 | NA | NA | NA | NA | 532618690 | NA | BWC08/02 | IN0802 | NA | f | caribou |
| BWC08.05 | NA | NA | NA | NA | 532618693 | NA | BWC08/05 | IN0805 | NA | f | caribou |
| BWC08.07 | NA | NA | NA | NA | 532618752 | NA | BWC08/07 | IN0807 | NA | f | caribou |
| BWC08.08 | NA | NA | NA | NA | 532618696 | NA | BWC08/08 | IN0808 | NA | f | caribou |
| BWC08.09 | NA | NA | NA | NA | 532618700 | NA | BWC08/09 | IN0809 | NA | f | caribou |
| BWC08.10 | NA | NA | NA | NA | 532618703 | NA | BWC08/10 | IN0810 | NA | f | caribou |
| BWC08.11 | NA | NA | NA | NA | 532618706 | NA | BWC08/11 | IN0811 | NA | f | caribou |
| BWC18 | NA | NA | NA | NA | 532618755 | NA | BWC18 | IN18 | NA | f | caribou |
| BWC21 | NA | NA | NA | NA | 532618709 | NA | BWC21 | IN21 | NA | f | caribou |
| BWC23 | NA | NA | NA | NA | 532618758 | NA | BWC23 | IN23 | NA | f | caribou |
| BWC35981 | NA | NA | NA | NA | 532618761 | NA | BWC35981 | IN35981 | NA | f | caribou |
| BWC35982 | NA | NA | NA | NA | 532618713 | NA | BWC35982 | IN35982 | NA | f | caribou |
| BWC35983 | NA | NA | NA | NA | 532618762 | NA | BWC35983 | IN35983 | NA | f | caribou |
| BWC35984 | NA | NA | NA | NA | 532618721 | NA | BWC35984 | IN35984 | NA | f | caribou |
| BWC36182 | NA | NA | NA | NA | 532618764 | NA | BWC36182 | IN36182 | NA | f | caribou |
| BWC36186 | NA | NA | NA | NA | 532622209 | NA | BWC36186 | IN36186 | NA | f | caribou |
| BWC36187 | NA | NA | NA | NA | 532622214 | NA | BWC36187 | IN36187 | NA | f | caribou |
| BWC36188 | NA | NA | NA | NA | 532622215 | NA | BWC36188 | IN36188 | NA | f | caribou |
| BWC36189 | NA | NA | NA | NA | 532622218 | NA | BWC36189 | IN36189 | NA | f | caribou |
Unfortunately, none of the columns in the @idData data frame actually corresponds to a column in the @data frame. However, the local_identifier column DOES correspond to another slot in the move object: the trackId:
table(inuvik@trackId)##
## BW26 BW27 BW28 BW29 BW30 BW31 BW32 BW33
## 4471 2122 2138 2101 2111 2127 5 229
## BW34 BW35 BW36 BW37 BW39 BW40 BW41 BW42
## 5519 139 139 288 509 732 456 565
## BW43 BW44 BW45 BWC07.01 BWC07.04 BWC07.05 BWC07.09 BWC08.01
## 15 4 3608 3527 2470 3604 1145 3208
## BWC08.02 BWC08.05 BWC08.07 BWC08.08 BWC08.09 BWC08.10 BWC08.11 BWC18
## 1265 1619 442 3519 2907 3231 3028 116
## BWC21 BWC23 BWC35981 BWC35982 BWC35983 BWC35984 BWC36182 BWC36186
## 4051 319 1303 1562 1164 1365 1996 275
## BWC36187 BWC36188 BWC36189
## 70 257 276This is a strange feature in move objects which I just don’t understand … why not have a consistently names column in both data frames?
2.3 Converting to a dataframe
The move package uses the trackId to knows which animal is which. When you convert the move object to a data frame (using the as.data.frame command), all of the information from the the id table carries over:
inuvik.df <- as.data.frame(inuvik)
names(inuvik.df)## [1] "sensor_type_id" "comments"
## [3] "habitat" "import_marked_outlier"
## [5] "location_lat" "location_long"
## [7] "manually_marked_outlier" "timestamp"
## [9] "update_ts" "argos_calcul_freq"
## [11] "argos_lc" "deployment_id"
## [13] "event_id" "tag_id"
## [15] "sensor_type" "location_long.1"
## [17] "location_lat.1" "optional"
## [19] "sensor" "timestamps"
## [21] "trackId" "comments.1"
## [23] "death_comments" "earliest_date_born"
## [25] "exact_date_of_birth" "individual_id"
## [27] "latest_date_born" "local_identifier"
## [29] "nick_name" "ring_id"
## [31] "sex" "taxon_canonical_name"Note that now we have the trackId, the tag_id, and lots of other ID’s, so we can cross-reference if needed:
inuvik.df[,c("trackId", "local_identifier", "nick_name", "deployment_id", "tag_id","individual_id")] %>% unique## trackId local_identifier nick_name deployment_id tag_id
## 25915 BW26 BW26 IN26 532845469 532618725
## 28068 BW26 BW26 IN26 532845476 532618727
## 38884 BW27 BW27 IN27 532845495 532618743
## 32523 BW28 BW28 IN28 532845510 532618734
## 34663 BW29 BW29 IN29 532845472 532618737
## 36768 BW30 BW30 IN30 532845483 532618740
## 30390 BW31 BW31 IN31 532845474 532618730
## 66339 BW32 BW32 IN32 532845505 532622171
## 66345 BW33 BW33 IN33 532845499 532622186
## 41014 BW34 BW34 IN34 532845473 532618747
## 43005 BW34 BW34 IN34 532845485 532618767
## 66574 BW35 BW35 IN35 532845486 532622189
## 66713 BW36 BW36 IN36 532845479 532622192
## 66852 BW37 BW37 IN37 532845506 532622195
## 67140 BW39 BW39 IN39 532845471 532622198
## 67649 BW40 BW40 IN40 532845496 532622201
## 68382 BW41 BW41 IN41 532845484 532622204
## 68838 BW42 BW42 IN42 532845468 532622207
## 46547 BW43 BW43 IN43 532845509 532618750
## 66335 BW44 BW44 IN44 532845501 532618800
## 51917 BW45 BW45 IN45 532845487 532839235
## 55525 BWC07.01 BWC07/01 IN0701 532845497 532618771
## 59081 BWC07.04 BWC07/04 IN0704 532845478 532618774
## 61566 BWC07.05 BWC07/05 IN0705 532845467 532618785
## 65185 BWC07.09 BWC07/09 IN0709 532845480 532618791
## 2 BWC08.01 BWC08/01 IN0801 532845511 532618681
## 3221 BWC08.02 BWC08/02 IN0802 532845491 532618691
## 4495 BWC08.05 BWC08/05 IN0805 532845470 532618694
## 46566 BWC08.07 BWC08/07 IN0807 532845477 532618753
## 6126 BWC08.08 BWC08/08 IN0808 532845488 532618697
## 9660 BWC08.09 BWC08/09 IN0809 532845503 532618701
## 12578 BWC08.10 BWC08/10 IN0810 532845507 532618704
## 15826 BWC08.11 BWC08/11 IN0811 532845490 532618707
## 47011 BWC18 BWC18 IN18 532845475 532618756
## 18875 BWC21 BWC21 IN21 532845492 532618710
## 47129 BWC23 BWC23 IN23 532845504 532618759
## 47448 BWC35981 BWC35981 IN35981 532845493 532618743
## 22955 BWC35982 BWC35982 IN35982 532845489 532618714
## 48754 BWC35983 BWC35983 IN35983 532845508 532618730
## 24533 BWC35984 BWC35984 IN35984 532845482 532618722
## 49918 BWC36182 BWC36182 IN36182 532845502 532618747
## 69403 BWC36186 BWC36186 IN36186 532845498 532622210
## 69678 BWC36187 BWC36187 IN36187 532845500 532622192
## 69748 BWC36188 BWC36188 IN36188 532845494 532622216
## 70005 BWC36189 BWC36189 IN36189 532845481 532622219
## individual_id
## 25915 532618724
## 28068 532618724
## 38884 532618742
## 32523 532618733
## 34663 532618736
## 36768 532618739
## 30390 532618729
## 66339 532622170
## 66345 532622185
## 41014 532618746
## 43005 532618746
## 66574 532622188
## 66713 532622191
## 66852 532622194
## 67140 532622197
## 67649 532622200
## 68382 532622203
## 68838 532622206
## 46547 532618749
## 66335 532618799
## 51917 532618765
## 55525 532618770
## 59081 532618773
## 61566 532618784
## 65185 532618790
## 2 532618680
## 3221 532618690
## 4495 532618693
## 46566 532618752
## 6126 532618696
## 9660 532618700
## 12578 532618703
## 15826 532618706
## 47011 532618755
## 18875 532618709
## 47129 532618758
## 47448 532618761
## 22955 532618713
## 48754 532618762
## 24533 532618721
## 49918 532618764
## 69403 532622209
## 69678 532622214
## 69748 532622215
## 70005 532622218Note that animal BW26 has, for example, multiple deployments.
3 Merging with external ID data
3.1 Ancillary data
For this population (and, apparently, many of the NWT woodland populations), there is some individual specific ancillary data avaialble in a an external file:
inuvik.ref.data <- read.csv("./_data/ABoVE_ NWT Inuvik Boreal Woodland Caribou-reference-data.csv")
names(inuvik.ref.data)## [1] "ï..tag.id" "animal.id"
## [3] "animal.taxon" "deploy.on.date"
## [5] "deploy.off.date" "animal.sex"
## [7] "animal.taxon.detail" "deploy.off.latitude"
## [9] "deploy.off.longitude" "deploy.on.latitude"
## [11] "deploy.on.longitude" "deployment.end.comments"
## [13] "deployment.end.type" "manipulation.type"
## [15] "study.site" "tag.beacon.frequency"
## [17] "tag.manufacturer.name" "tag.model"
## [19] "tag.readout.method"Note - that bizarro character ?.. in front of the first column is some strange Unicode encoding issue (which may or may not show up when you try to replicate this). We have to reload the data with an extra encoding option to remove it:
Note, also, that the original data file had columns that used dashes as separators, e.g.: tag-id, animal-id, animal-taxon, deploy-on-date, deploy-off-date, etc. These are transformed into periods when read into R, since the dash is a minus sign.
This data frame contains several many columns that aren’t in the inuvik@idData slot, compare:
names(inuvik@idData)## [1] "comments" "death_comments" "earliest_date_born"
## [4] "exact_date_of_birth" "individual_id" "latest_date_born"
## [7] "local_identifier" "nick_name" "ring_id"
## [10] "sex" "taxon_canonical_name"3.2 Merging with idData
We can merge all of the information in the reference data frame using the very powerful merge function. Note that you have to specify the columns by which you’re matching the data frames, in this case: local_identifier matches the tag.id. Note that we use all.x = TRUE, which retains only rows of the first data frame, but not necessarily all the rows of the second data frame, in order to to keep the same basic shape as the original @idData.
inuvik.idData.merged <- merge(inuvik@idData, inuvik.ref.data,
by.x = "local_identifier",
by.y = "animal.id",
all.x = TRUE)This created a merged data frame, with, however, slightly different number of rows than @idData:
dim(inuvik.idData.merged)## [1] 45 29dim(inuvik@idData)## [1] 43 11This is no good! It happens because there are a few “animal.id”’s in the refrence data with multiple rows. Here’s some quick code to see which ones:
tail(sort(table(inuvik.ref.data$animal.id)))##
## BWC36186 BWC36187 BWC36188 BWC36189 BW26 BW34
## 1 1 1 1 2 2I’m not sure why there are multiple rows, but since they are associated with only a single row in the
inuvikreference data, there might be some confusion! We need to eliminate the excess rows for the following to work.
which(inuvik.ref.data$animal.id == "BW26")## [1] 10 29which(inuvik.ref.data$animal.id == "BW34")## [1] 16 30inuvik.ref.data <- inuvik.ref.data[,-c(29,30)]
inuvik.idData.merged <- merge(inuvik@idData, inuvik.ref.data,
by.x = "local_identifier",
by.y = "animal.id",
all.x = TRUE)Unfortunately, we can’t simply replace the @idData with this new data frame. When we do so, and convert to a data frame all the information is lost. See:
inuvik2 <- inuvik
inuvik2@idData <- inuvik.idData.merged
as.data.frame(inuvik2) %>% str## 'data.frame': 69997 obs. of 50 variables:
## $ sensor_type_id : int 653 653 653 653 653 653 653 653 653 653 ...
## $ comments : logi NA NA NA NA NA NA ...
## $ habitat : logi NA NA NA NA NA NA ...
## $ import_marked_outlier : logi NA NA NA NA NA NA ...
## $ location_lat : num 67.5 67.5 67.5 67.5 67.5 ...
## $ location_long : num -132 -132 -132 -132 -132 ...
## $ manually_marked_outlier: Factor w/ 2 levels "false","true": 1 1 1 1 1 1 1 1 1 1 ...
## $ timestamp : POSIXct, format: "2005-03-11 00:00:00" "2005-03-11 08:03:42" ...
## $ update_ts : Factor w/ 2 levels "2018-09-17 20:32:20.291",..: 2 2 2 2 2 2 2 2 2 2 ...
## $ argos_calcul_freq : num NA NA NA NA NA NA NA NA NA NA ...
## $ argos_lc : int NA NA NA NA NA NA NA NA NA NA ...
## $ deployment_id : int 532845469 532845469 532845469 532845469 532845469 532845469 532845469 532845469 532845469 532845469 ...
## $ event_id : num 7.41e+09 7.41e+09 7.41e+09 7.41e+09 7.41e+09 ...
## $ tag_id : int 532618725 532618725 532618725 532618725 532618725 532618725 532618725 532618725 532618725 532618725 ...
## $ sensor_type : Factor w/ 2 levels "Argos Doppler Shift",..: 2 2 2 2 2 2 2 2 2 2 ...
## $ location_long.1 : num -132 -132 -132 -132 -132 ...
## $ location_lat.1 : num 67.5 67.5 67.5 67.5 67.5 ...
## $ optional : logi TRUE TRUE TRUE TRUE TRUE TRUE ...
## $ sensor : Factor w/ 2 levels "Argos Doppler Shift",..: 2 2 2 2 2 2 2 2 2 2 ...
## $ timestamps : POSIXct, format: "2005-03-11 00:00:00" "2005-03-11 08:03:42" ...
## $ trackId : Factor w/ 43 levels "BW26","BW27",..: 1 1 1 1 1 1 1 1 1 1 ...
## $ local_identifier : Factor w/ 43 levels "BW26","BW27",..: NA NA NA NA NA NA NA NA NA NA ...
## $ comments.1 : logi NA NA NA NA NA NA ...
## $ death_comments : logi NA NA NA NA NA NA ...
## $ earliest_date_born : logi NA NA NA NA NA NA ...
## $ exact_date_of_birth : logi NA NA NA NA NA NA ...
## $ individual_id : int NA NA NA NA NA NA NA NA NA NA ...
## $ latest_date_born : logi NA NA NA NA NA NA ...
## $ nick_name : Factor w/ 43 levels "IN0701","IN0704",..: NA NA NA NA NA NA NA NA NA NA ...
## $ ring_id : logi NA NA NA NA NA NA ...
## $ sex : Factor w/ 1 level "f": NA NA NA NA NA NA NA NA NA NA ...
## $ taxon_canonical_name : Factor w/ 1 level "caribou": NA NA NA NA NA NA NA NA NA NA ...
## $ ï..tag.id : Factor w/ 41 levels "12179","12181",..: NA NA NA NA NA NA NA NA NA NA ...
## $ animal.taxon : Factor w/ 1 level "Rangifer tarandus": NA NA NA NA NA NA NA NA NA NA ...
## $ deploy.on.date : Factor w/ 13 levels "2002-05-01 00:00:00.000",..: NA NA NA NA NA NA NA NA NA NA ...
## $ deploy.off.date : Factor w/ 33 levels "2003-10-27 23:59:00.000",..: NA NA NA NA NA NA NA NA NA NA ...
## $ animal.sex : Factor w/ 1 level "f": NA NA NA NA NA NA NA NA NA NA ...
## $ animal.taxon.detail : Factor w/ 1 level "caribou": NA NA NA NA NA NA NA NA NA NA ...
## $ deploy.off.latitude : num NA NA NA NA NA NA NA NA NA NA ...
## $ deploy.off.longitude : num NA NA NA NA NA NA NA NA NA NA ...
## $ deploy.on.latitude : num NA NA NA NA NA NA NA NA NA NA ...
## $ deploy.on.longitude : num NA NA NA NA NA NA NA NA NA NA ...
## $ deployment.end.comments: Factor w/ 25 levels "can be stationary much before 1/10/2006, not quality data, mulfunction afer 5/8/2006",..: NA NA NA NA NA NA NA NA NA NA ...
## $ deployment.end.type : Factor w/ 5 levels "dead","equipment-failure",..: NA NA NA NA NA NA NA NA NA NA ...
## $ manipulation.type : Factor w/ 1 level "none": NA NA NA NA NA NA NA NA NA NA ...
## $ study.site : Factor w/ 1 level "Inuvik": NA NA NA NA NA NA NA NA NA NA ...
## $ tag.beacon.frequency : num NA NA NA NA NA NA NA NA NA NA ...
## $ tag.manufacturer.name : Factor w/ 1 level "Telonics": NA NA NA NA NA NA NA NA NA NA ...
## $ tag.model : Factor w/ 3 levels "ST-10","ST-14",..: NA NA NA NA NA NA NA NA NA NA ...
## $ tag.readout.method : Factor w/ 1 level "satellite": NA NA NA NA NA NA NA NA NA NA ...All the new data is NA!?
3.3 Merging with movement data frame
So we have to “manually” merge the location data frame with the ORIGINAL reference data frame, using the trackID as the merging criterion. Thus:
inuvik.df <- as.data.frame(inuvik)
inuvik.df <- merge(inuvik.df, inuvik.ref.data,
by.x = "trackId", by.y = "animal.id", all.x = TRUE)
str(inuvik.df)## 'data.frame': 79987 obs. of 50 variables:
## $ trackId : Factor w/ 43 levels "BW26","BW27",..: 1 1 1 1 1 1 1 1 1 1 ...
## $ sensor_type_id : int 653 653 653 653 653 653 653 653 653 653 ...
## $ comments : logi NA NA NA NA NA NA ...
## $ habitat : logi NA NA NA NA NA NA ...
## $ import_marked_outlier : logi NA NA NA NA NA NA ...
## $ location_lat : num 68.3 68.3 68.3 68.3 68.3 ...
## $ location_long : num -133 -133 -133 -133 -133 ...
## $ manually_marked_outlier: Factor w/ 2 levels "false","true": 1 1 1 1 1 1 1 1 1 1 ...
## $ timestamp : POSIXct, format: "2005-07-18 16:01:55" "2005-07-18 16:01:55" ...
## $ update_ts : Factor w/ 2 levels "2018-09-17 20:32:20.291",..: 2 2 2 2 2 2 2 2 2 2 ...
## $ argos_calcul_freq : num NA NA NA NA NA NA NA NA NA NA ...
## $ argos_lc : int NA NA NA NA NA NA NA NA NA NA ...
## $ deployment_id : int 532845469 532845469 532845469 532845469 532845469 532845469 532845469 532845469 532845469 532845469 ...
## $ event_id : num 7.41e+09 7.41e+09 7.41e+09 7.41e+09 7.41e+09 ...
## $ tag_id : int 532618725 532618725 532618725 532618725 532618725 532618725 532618725 532618725 532618725 532618725 ...
## $ sensor_type : Factor w/ 2 levels "Argos Doppler Shift",..: 2 2 2 2 2 2 2 2 2 2 ...
## $ location_long.1 : num -133 -133 -133 -133 -133 ...
## $ location_lat.1 : num 68.3 68.3 68.3 68.3 68.3 ...
## $ optional : logi TRUE TRUE TRUE TRUE TRUE TRUE ...
## $ sensor : Factor w/ 2 levels "Argos Doppler Shift",..: 2 2 2 2 2 2 2 2 2 2 ...
## $ timestamps : POSIXct, format: "2005-07-18 16:01:55" "2005-07-18 16:01:55" ...
## $ comments.1 : logi NA NA NA NA NA NA ...
## $ death_comments : logi NA NA NA NA NA NA ...
## $ earliest_date_born : logi NA NA NA NA NA NA ...
## $ exact_date_of_birth : logi NA NA NA NA NA NA ...
## $ individual_id : int 532618724 532618724 532618724 532618724 532618724 532618724 532618724 532618724 532618724 532618724 ...
## $ latest_date_born : logi NA NA NA NA NA NA ...
## $ local_identifier : Factor w/ 43 levels "BW26","BW27",..: 1 1 1 1 1 1 1 1 1 1 ...
## $ nick_name : Factor w/ 43 levels "IN0701","IN0704",..: 16 16 16 16 16 16 16 16 16 16 ...
## $ ring_id : logi NA NA NA NA NA NA ...
## $ sex : Factor w/ 1 level "f": 1 1 1 1 1 1 1 1 1 1 ...
## $ taxon_canonical_name : Factor w/ 1 level "caribou": 1 1 1 1 1 1 1 1 1 1 ...
## $ ï..tag.id : Factor w/ 41 levels "12179","12181",..: 26 2 26 2 26 2 26 2 26 2 ...
## $ animal.taxon : Factor w/ 1 level "Rangifer tarandus": 1 1 1 1 1 1 1 1 1 1 ...
## $ deploy.on.date : Factor w/ 13 levels "2002-05-01 00:00:00.000",..: 8 4 8 4 8 4 8 4 8 4 ...
## $ deploy.off.date : Factor w/ 33 levels "2003-10-27 23:59:00.000",..: 26 14 26 14 26 14 26 14 26 14 ...
## $ animal.sex : Factor w/ 1 level "f": 1 1 1 1 1 1 1 1 1 1 ...
## $ animal.taxon.detail : Factor w/ 1 level "caribou": 1 1 1 1 1 1 1 1 1 1 ...
## $ deploy.off.latitude : num 67.6 67.7 67.6 67.7 67.6 ...
## $ deploy.off.longitude : num -132 -132 -132 -132 -132 ...
## $ deploy.on.latitude : num 67.7 67.6 67.7 67.6 67.7 ...
## $ deploy.on.longitude : num -132 132 -132 132 -132 ...
## $ deployment.end.comments: Factor w/ 25 levels "can be stationary much before 1/10/2006, not quality data, mulfunction afer 5/8/2006",..: 11 8 11 8 11 8 11 8 11 8 ...
## $ deployment.end.type : Factor w/ 5 levels "dead","equipment-failure",..: 1 4 1 4 1 4 1 4 1 4 ...
## $ manipulation.type : Factor w/ 1 level "none": 1 1 1 1 1 1 1 1 1 1 ...
## $ study.site : Factor w/ 1 level "Inuvik": 1 1 1 1 1 1 1 1 1 1 ...
## $ tag.beacon.frequency : num 150 151 150 151 150 ...
## $ tag.manufacturer.name : Factor w/ 1 level "Telonics": 1 1 1 1 1 1 1 1 1 1 ...
## $ tag.model : Factor w/ 3 levels "ST-10","ST-14",..: 3 3 3 3 3 3 3 3 3 3 ...
## $ tag.readout.method : Factor w/ 1 level "satellite": 1 1 1 1 1 1 1 1 1 1 ...We now have ALL that ancillary data attached to the inuvik.df data frame.
Note, this is a BIG data frame! As a general R workflow recommendation, it is a good idea to save this object in R’s native format to quickly reload it as needed, e.g.:
save(inuvik.df, file = "./_data/InuvikProcessed.rda")So that when you revisit the code you can quickly load the data into your namespace via:
load("./_data/InuvikProcessed.rda")4 Plotting the data
Ok, enough messing with data. Let’s do some plotting! Note, we can load our processed data to skip a lot of the steps above:
load("./_data/InuvikProcessed.rda")4.1 Basic plot
A useful tool for plotting structured data like this (x-y-id) is with ggplot. There are lots of references online, but here is an example:
require(ggplot2)
ggplot(inuvik.df, aes(location_long, location_lat, col = nick_name)) + geom_path() + geom_point()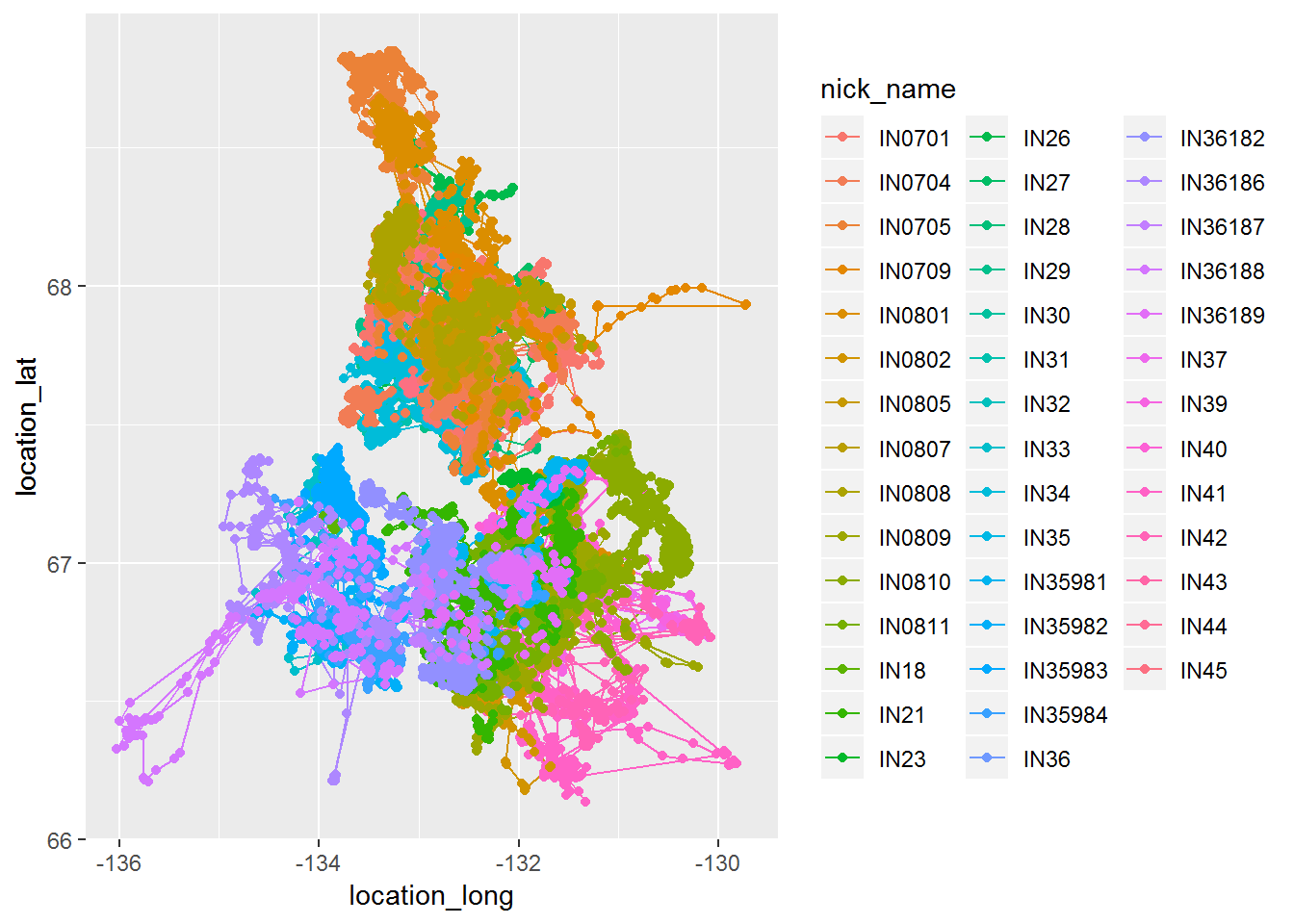
4.2 GGmaps
There is a package called ggmap with allows us to use Google and other available map sources to improve our plots. There are several steps here:
load(file = "./_data/inuvikmap.rda")- Load
ggmap(or first install it if you don’t have it) and obtain a “basemap”, which requires a range and a zoom level.
require(ggmap)
latrange <- range(inuvik.df$location_lat) + c(-.5, .5)
longrange <- range(inuvik.df$location_long) + c(-.5, .5)
## inuvik.map <- get_map(location = c(longrange[1], latrange[1], longrange[2], latrange[2]), maptype = "terrain", source = "google")There are lots of options for the maptype, e.g. “terrain”, “satellite”, “roadmap”, “hybrid”. The choice depends on the information you want to communicate (and how much you want to foreground your tracks).
Note: if your data is a Move object, you can use the
bbox()command to automatically obtain the bounding box of the data range code as follows:
require(move)
inuvik.map <- get_map(bbox(inuvik), source="google", maptype="satellite")- There will be no output - you just created an objet called
inuvik.map, which you can view with theggmapcommand:
ggmap(inuvik.map)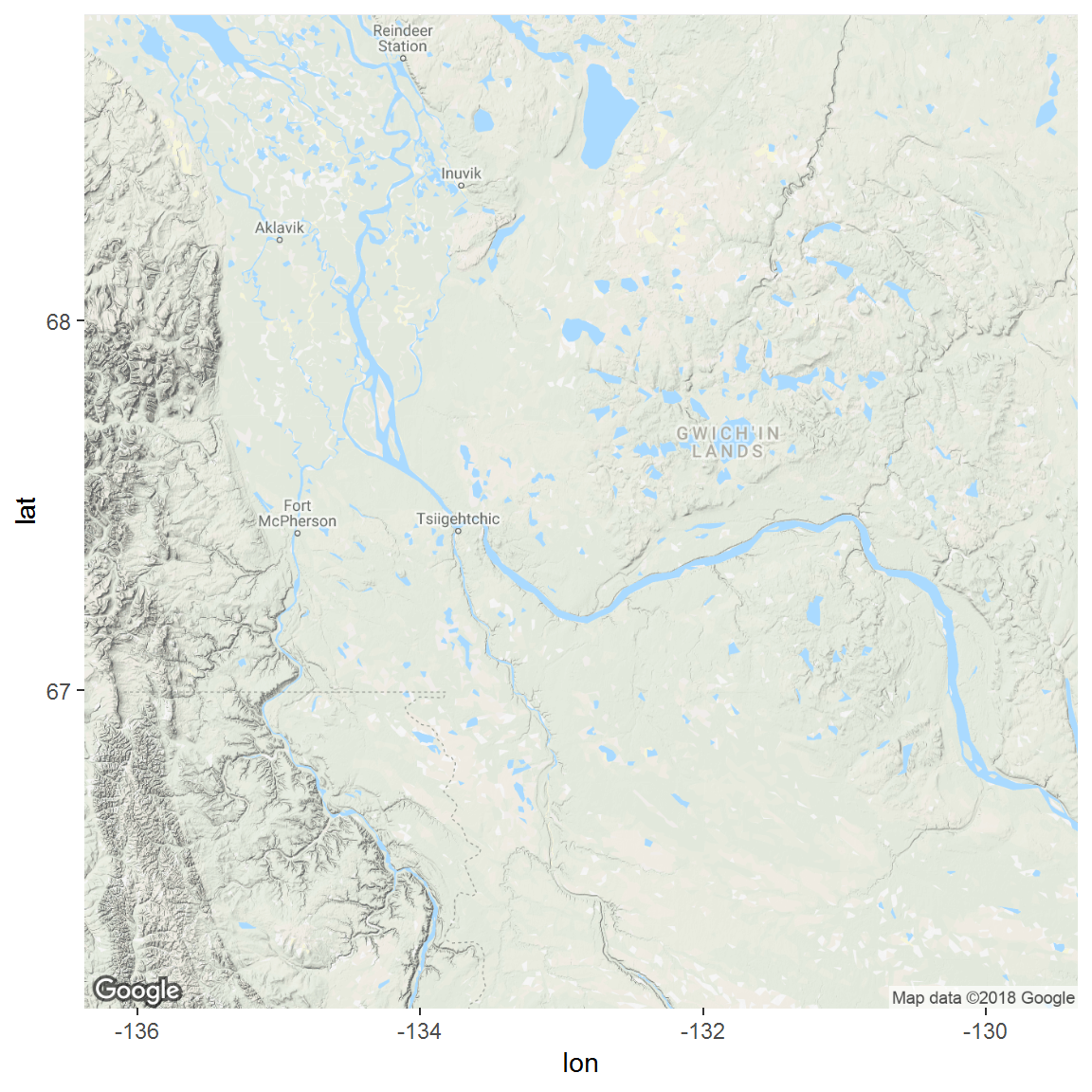
then add your data onto that map
ggmap(inuvik.map) +
geom_point(data = inuvik.df, mapping = aes(x = location_long, y = location_lat, col = nick_name), alpha = 0.5, size = 0.5) +
geom_path(data = inuvik.df, mapping = aes(x = location_long, y = location_lat, col = nick_name), alpha = 0.5, size = 0.5) +
labs(x="Longitude", y="Latitude", main = "Inuvik Caribou")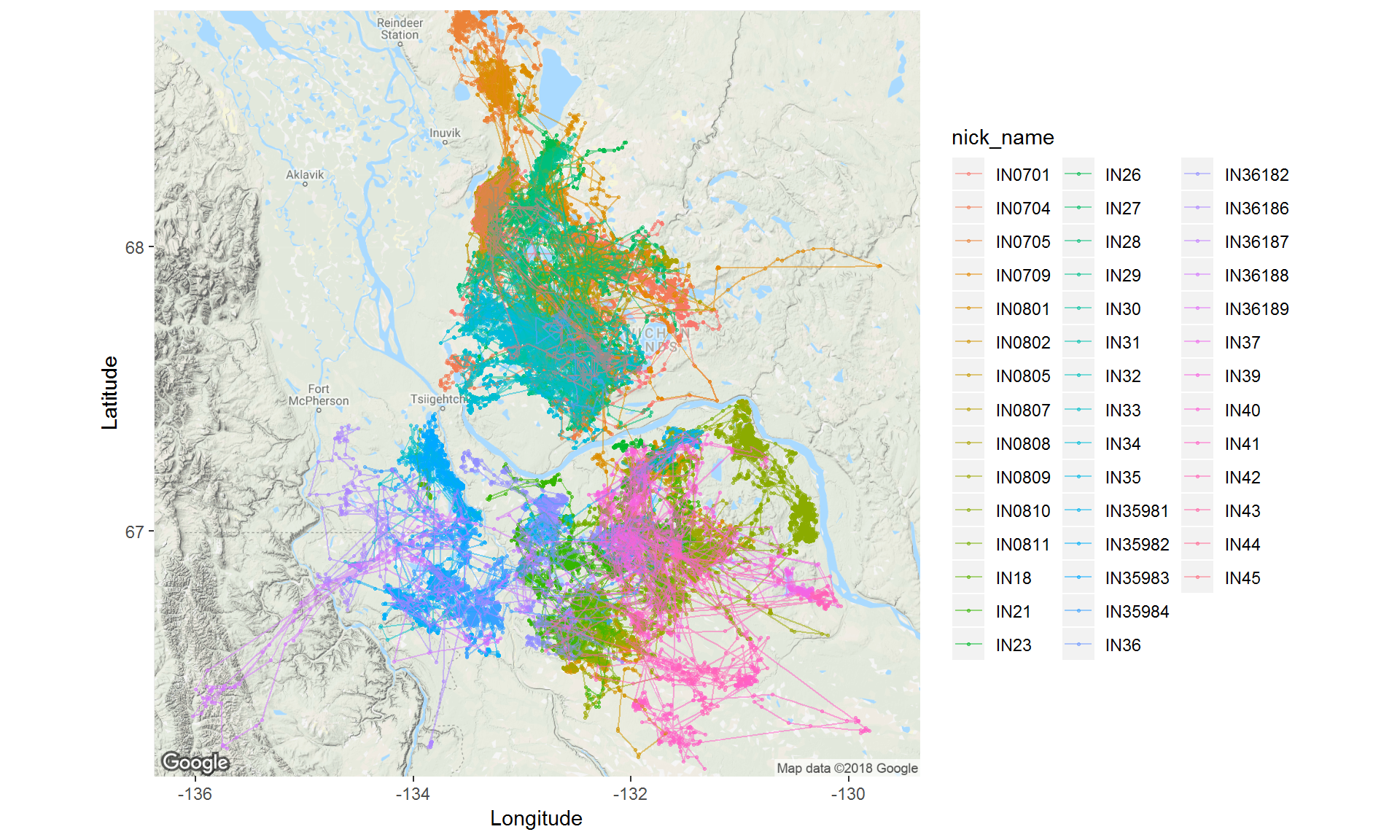
BAD NEWS (UPDATE OCTOBER 27, 2018): The code above is not guaranteed to work since Google has decided to make their maps API no longer (entirely) free. You can still do this, but you will need an “api_key”, for which you need to enable billing (see here: https://developers.google.com/maps/documentation/javascript/get-api-key). This is a major drag. However, there are other cool options …..
4.3 Mapview
There is a very powerful open-source scripting library called leaflet, which allows for the creation of interactive maps with a large range of backgrounds. It’s remarkable easy to use from R, but there is some prep work that needs to be done.
The key packages:
require(leaflet)
require(mapview) 4.3.1 First, simple features
We are going to convert our inuvik data into a special kind of object (to which we will be returning to often) called a simple feature, which is basically a data frame where one column contains all of the spatial information. It is easy to create a simple feature object out of a move object. Below we do the conversion, and merge it with some extra useful information from the inuvik.df:
require(sf)
inuvik.sf <- st_as_sf(inuvik)
inuvik.sf <- merge(inuvik.sf, inuvik.df[,c("timestamp","deployment_id","event_id","nick_name")])Simple features allow us to do some nicely projected mapping. For example:
ggplot(subset(inuvik.sf, nick_name == nick_name[1]), aes(location_long, location_lat, col = nick_name)) + geom_sf() + geom_path()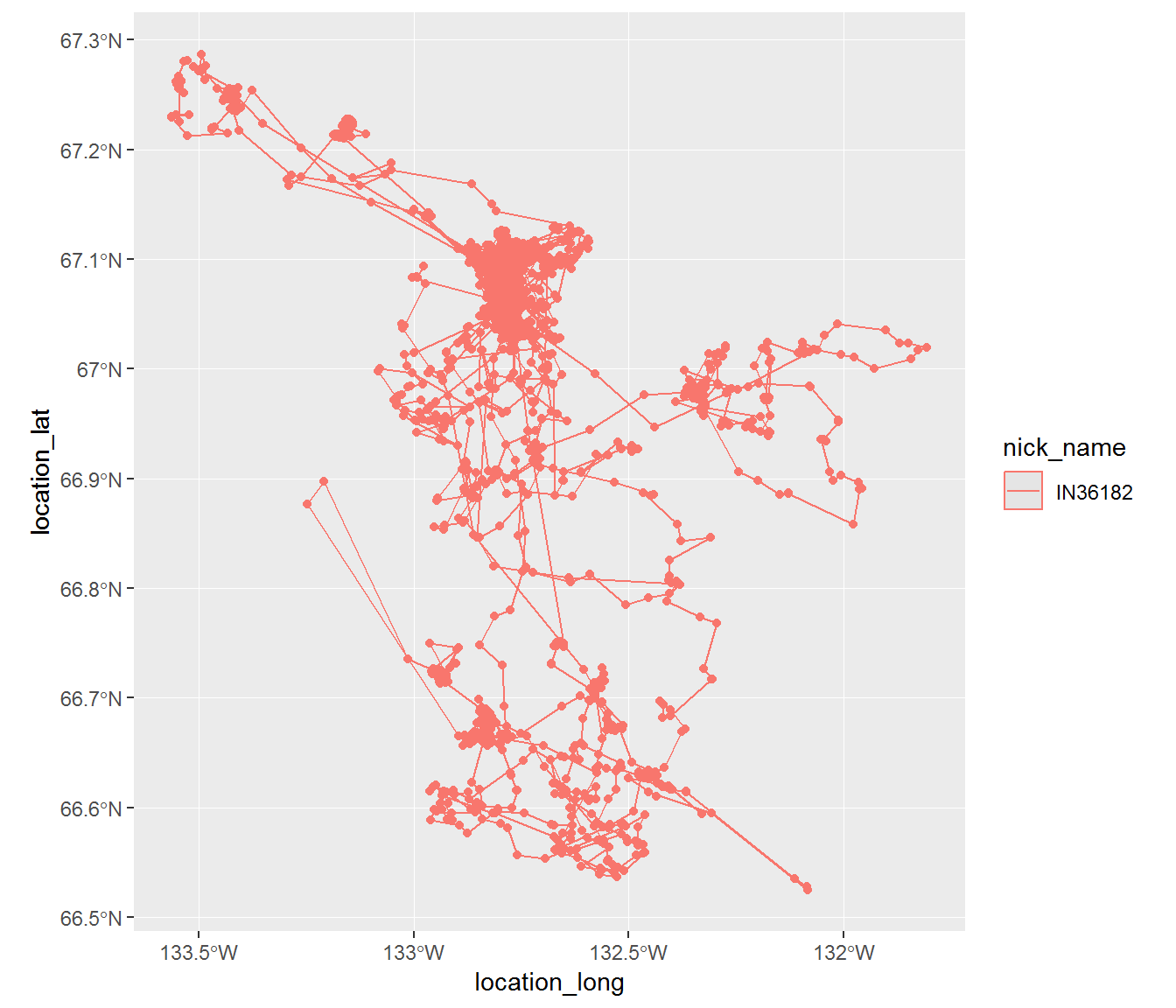
But if you wanted it projected to, e.g., Canada Albers Equal Area Conic (ESRI:102002)
ggplot(subset(inuvik.sf, nick_name == nick_name[1]) %>% st_transform(102002), aes(col = nick_name)) + geom_sf()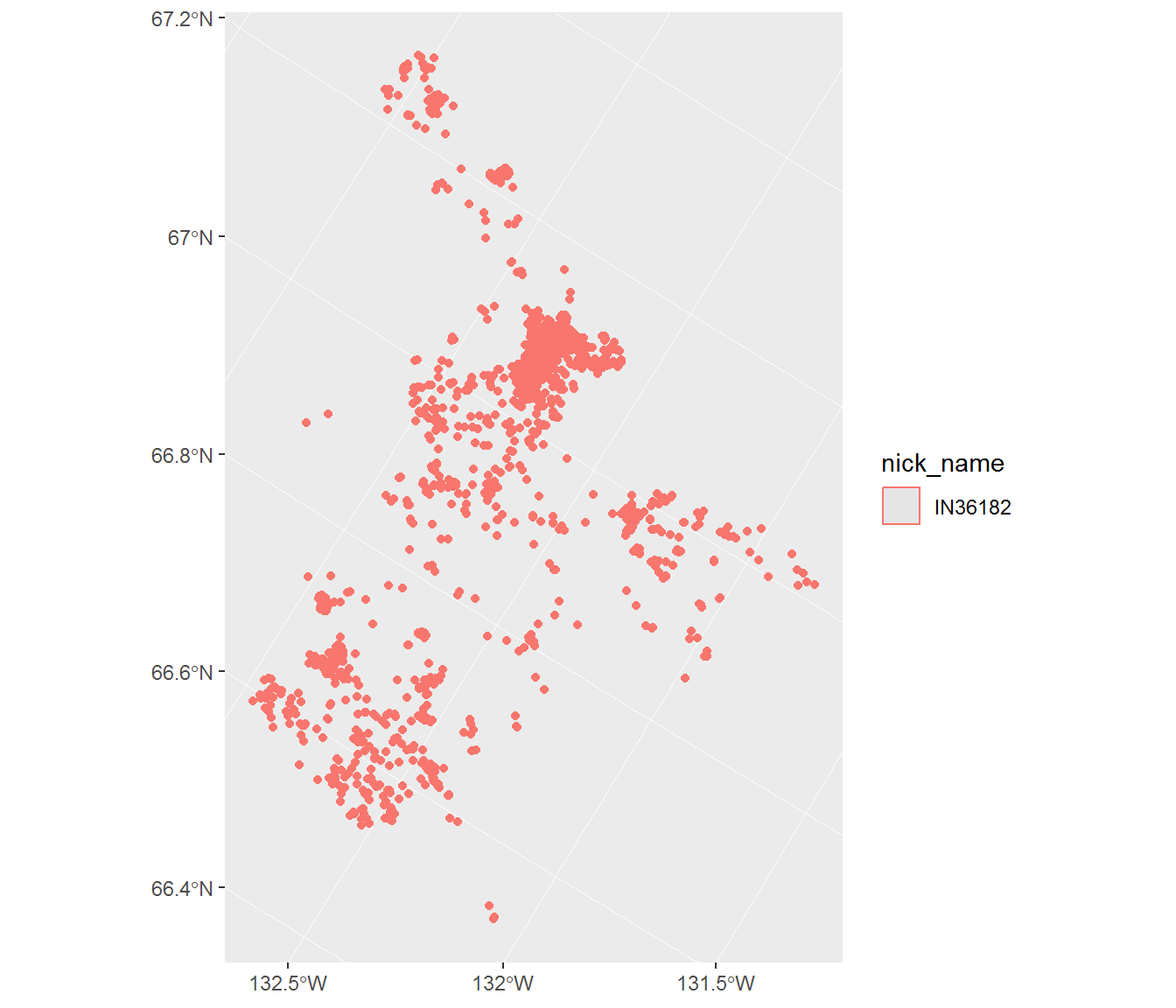
To turn the above into a path, which is what we really want, we need to “collapse” the POINT geometry of this caribou, into a single LINES geometry. That’s done as follows (and I confess I don’t entirely understand the sequence of dplyr commands, but it doesn’t seem to work without the grouping AND the summarizing):
require(dplyr)
inuvik.line <-inuvik.sf[,"nick_name"] %>% group_by(nick_name) %>% summarize(do_union=FALSE) %>% st_cast("LINESTRING")
ggplot(inuvik.line %>% st_transform(102002), aes(col = nick_name)) + geom_sf() + ggtitle("Inuvik barrenground caribou: Canada Lambert Conformal Conic")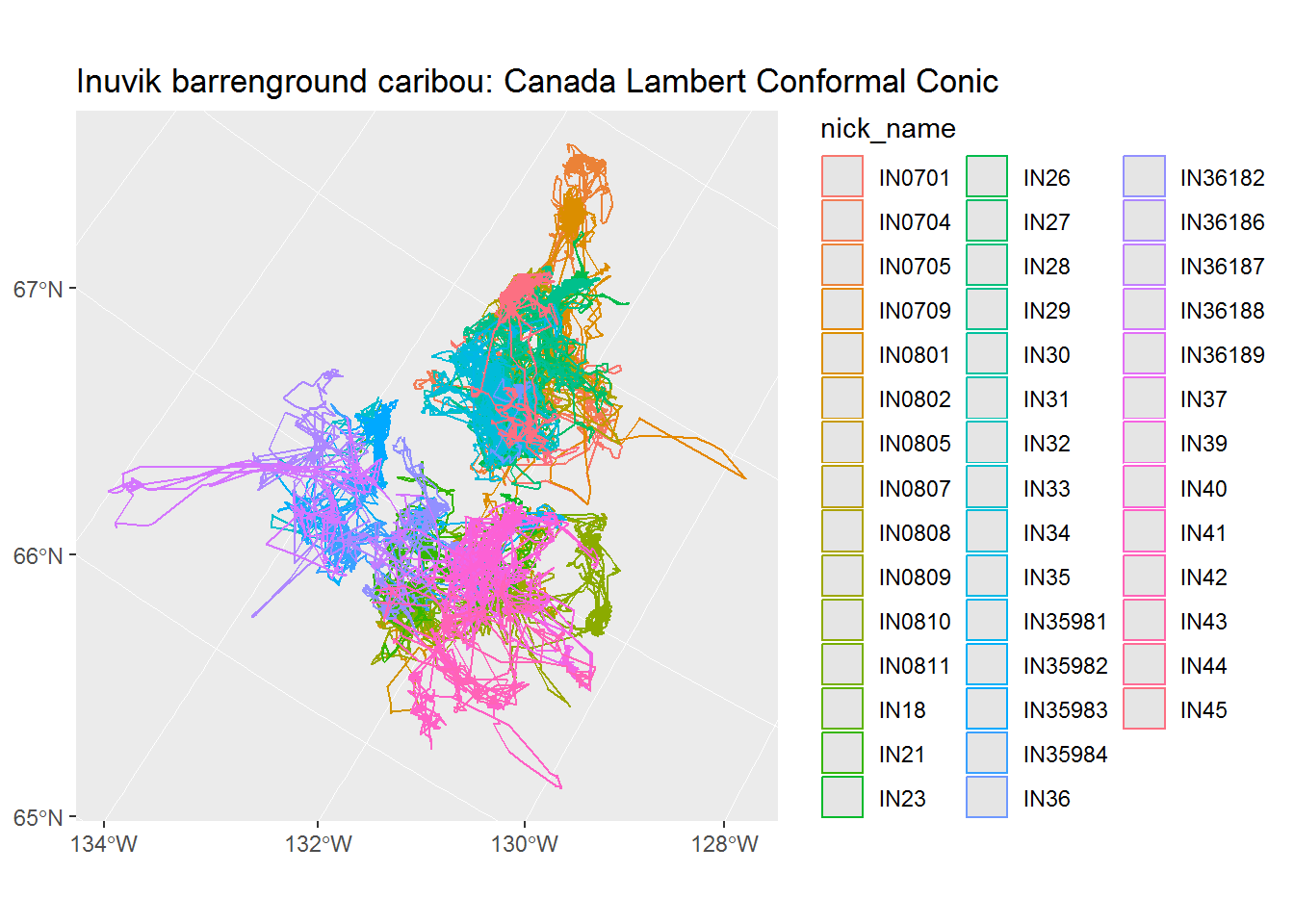
4.3.2 Interactive mapping
Now that the LINES simple feature data frame is set up, plotting this is as easy as:
mapview(inuvik.line, zcol = "nick_name", map.types = "Esri.WorldPhysical")Note: this is an interactive map. You can zoom it, drag it, and scroll over the paths a nd see the ID of the animal. “Esri.WorldPhysical” is just one of many possible maps. Here’s a link to a bunch of other ones. Here’s another, more stylized one:
mapview(inuvik.line, zcol = "nick_name", map.types = "Esri.NatGeoWorldMap")Easy! And quite useful, I think.
5 Complete code
Click below to show complete code from this document.
knitr::opts_chunk$set(echo = TRUE, cache = TRUE, warning = FALSE, message = FALSE)
pcks <- c("magrittr", "rmarkdown")
a <- lapply(pcks, require, character = TRUE)
require(move)
mylogin <- movebankLogin(username="Elie Gurarie", password="Caribou")
## mylogin <- movebankLogin(username="MyUserName", password="MyPassword")
inuvik <- getMovebankData("ABoVE: NWT Inuvik Boreal Woodland Caribou", login = mylogin)
## save(inuvik, file = "./_data/inuvik.rda")
load(file = "./_data/inuvik.rda")
inuvik
slotNames(inuvik)
## head(inuvik@data)
head(inuvik@data) %>% kable
inuvik@proj4string
inuvik@license
inuvik@citation
## inuvik@idData
require(knitr)
kable(inuvik@idData)
table(inuvik@trackId)
inuvik.df <- as.data.frame(inuvik)
names(inuvik.df)
inuvik.df[,c("trackId", "local_identifier", "nick_name", "deployment_id", "tag_id","individual_id")] %>% unique
## setwd("C:/Users/Guru/box sync/caribou/NWT/rsf/primers")
## load("./_data/inuvik.rda")
inuvik.ref.data <- read.csv("./_data/ABoVE_ NWT Inuvik Boreal Woodland Caribou-reference-data.csv")
names(inuvik.ref.data)
## inuvik.ref.data <- read.csv("./_data/ABoVE_ NWT Inuvik Boreal Woodland Caribou-reference-data.csv", fileEncoding="UTF-8-BOM")
## names(inuvik.ref.id)
names(inuvik@idData)
inuvik.idData.merged <- merge(inuvik@idData, inuvik.ref.data,
by.x = "local_identifier",
by.y = "animal.id",
all.x = TRUE)
dim(inuvik.idData.merged)
dim(inuvik@idData)
tail(sort(table(inuvik.ref.data$animal.id)))
which(inuvik.ref.data$animal.id == "BW26")
which(inuvik.ref.data$animal.id == "BW34")
inuvik.ref.data <- inuvik.ref.data[,-c(29,30)]
inuvik.idData.merged <- merge(inuvik@idData, inuvik.ref.data,
by.x = "local_identifier",
by.y = "animal.id",
all.x = TRUE)
inuvik2 <- inuvik
inuvik2@idData <- inuvik.idData.merged
as.data.frame(inuvik2) %>% str
inuvik.df <- as.data.frame(inuvik)
inuvik.df <- merge(inuvik.df, inuvik.ref.data,
by.x = "trackId", by.y = "animal.id", all.x = TRUE)
str(inuvik.df)
## save(inuvik.df, file = "./_data/InuvikProcessed.rda")
## load("./_data/InuvikProcessed.rda")
load("./_data/InuvikProcessed.rda")
require(ggplot2)
ggplot(inuvik.df, aes(location_long, location_lat, col = nick_name)) + geom_path() + geom_point()
load(file = "./_data/inuvikmap.rda")
require(ggmap)
latrange <- range(inuvik.df$location_lat) + c(-.5, .5)
longrange <- range(inuvik.df$location_long) + c(-.5, .5)
inuvik.map <- get_map(location = c(longrange[1], latrange[1], longrange[2], latrange[2]), maptype = "terrain", source = "google")
## save(inuvik.map, file = "./_data/inuvikmap.rda")
## require(move)
## inuvik.map <- get_map(bbox(inuvik), source="google", maptype="satellite")
ggmap(inuvik.map)
ggmap(inuvik.map) +
geom_point(data = inuvik.df, mapping = aes(x = location_long, y = location_lat, col = nick_name), alpha = 0.5, size = 0.5) +
geom_path(data = inuvik.df, mapping = aes(x = location_long, y = location_lat, col = nick_name), alpha = 0.5, size = 0.5) +
labs(x="Longitude", y="Latitude", main = "Inuvik Caribou")
require(leaflet)
require(mapview)
require(sf)
inuvik.sf <- st_as_sf(inuvik)
inuvik.sf <- merge(inuvik.sf, inuvik.df[,c("timestamp","deployment_id","event_id","nick_name")])
ggplot(subset(inuvik.sf, nick_name == nick_name[1]), aes(location_long, location_lat, col = nick_name)) + geom_sf() + geom_path()
ggplot(subset(inuvik.sf, nick_name == nick_name[1]) %>% st_transform(102002), aes(col = nick_name)) + geom_sf()
require(dplyr)
inuvik.line <-inuvik.sf[,"nick_name"] %>% group_by(nick_name) %>% summarize(do_union=FALSE) %>% st_cast("LINESTRING")
ggplot(inuvik.line %>% st_transform(102002), aes(col = nick_name)) + geom_sf() + ggtitle("Inuvik barrenground caribou: Canada Lambert Conformal Conic")
require(mapview)
mapview(inuvik.line, zcol = "nick_name", map.types = "Esri.WorldPhysical")
require(mapview)
mapview(inuvik.line, zcol = "nick_name", map.types = "Esri.NatGeoWorldMap")