Lab 9b: Bayesian Inference, the numerical approach
BIOL709/BSCI339
There are, essentially, three different ways to obtain Bayesian estimates of parameters: (a) analytically compute them; (b) numerically perform the updating of the prior across a fine mesh of possible values for the parameters; or (c) sample from the posterior with a Markov chain Monte Carlo.
The first option is possible for only a relatively narrow set of models with well-behaved conjugate prior-likelihood pairs.
The second - which we perform here - is more flexible, but often computationally prohibitive.
The third we will address in the final week of class.
Everything in this lab uses base R, but I often find it irresistible to do some piping!
require(magrittr)Revisiting the basketball player data
We’re going to obtain a Bayesian estimate of the mean and variance of (a small simulated set) of basketball player heights and compare it to the MLE. Here are some data:
mu <- 6.5
sigma <- 1
X <- rnorm(10, mu, sigma) %>% sort %>% round(2) %T>% print## [1] 4.80 5.42 5.50 6.32 6.62 6.73 7.17 7.41 8.12 8.80We are going to assume that the data are distributed normally and try to estimate the two parameters \(\mu\) and \(\sigma\), comparing the Bayesian estimate to our familiar MLE estimates.
Bayes’ theorem
The goal in any Bayesian analsyis is to obtain a posterior distribution from Bayes’ theorem which says: \[\pi(\mu, \sigma | X) \propto p(X|\mu, \sigma) \pi(\mu, \sigma).\] The steps are straightforward: (1) select priors, (2) define the likelihood, (3) multiply [i.e. integrate] the two functions for “all” possible values of the parameters.
Priors
Let’s use uninformative priors. For the mean, one possibility is a uniform distribution over the range of the data, since the mean must be somewhere in that range:
\[\mu \sim \text{Unif}(X_{min}, X_{min})\] \[ \pi(\mu) = {1 \over X_{max} - X_{min}}\] where \(X_{min} \leq \mu \leq X_{max}\).
For \(\sigma\), it is harder to place bounds, but a nice uninformative positive prior can be the exponential:
\[\sigma \sim \text{Exp}(\lambda_0)\] \[\pi(\sigma) = \lambda \exp(-\lambda \sigma)\] where \(\sigma > 0\) and \(\lambda\) is some very small number.
Set these parameters for the priors:
mu.min <- min(X)
mu.max <- max(X)
lambda <- 1e-4Likelihood function
The likelihood function (as in all of our estimations) is our actual data model, i.e. the normal distribution: \[p({\bf X}|\mu, \sigma) = \prod_{i=1}^n \phi(X_i, \mu, \sigma)\] wher \(\phi\) is the p.d.f. of the normal distribution.
likelihood <- function(x, mu, sigma) prod(dnorm(X, mu, sigma))Note that we can not be in the “log-likelihood” world which we prefer for maximum likelihood estimation, since we need the actual probability distribution. That said, for larger amounts of data you would need to toggle back and forth between the log and untransformed units, e.g.:
likelihood <- function(x, mu, sigma) exp(sum(dnorm(X, mu, sigma, log=TRUE)))Integration
Now that we have all the pieces, we need to compute the posterior distribution: \(\pi(\mu, \sigma)\), which is (proportional) simply to the product of the priors and the likelihood at every value of \(\mu\) and \(\sigma\):
getposterior <- function(mu, sigma)
likelihood(X, mu, sigma)*dunif(mu, mu.min, mu.max)*dexp(sigma, lambda)Initialize vectors for the two parameters (at reasonably high resolution):
dmu <- .01; dsigma <- .01
mus <- seq(min(X),max(X), dmu)
sigmas <- seq(dsigma,5, dsigma)The slickest way to perform the calculation of the posteriors for all values is our friend outer():
posterior <- outer(mus, sigmas, Vectorize(getposterior))NOTE: this posterior is not normalized! How would we normalize it if we wanted to?
Here is a visualization of the posterior density:
image(mus, sigmas, posterior, ylim=c(0,2), col=topo.colors(200))
persp(mus, sigmas, posterior, border=NA, shade=TRUE, ticktype="detailed")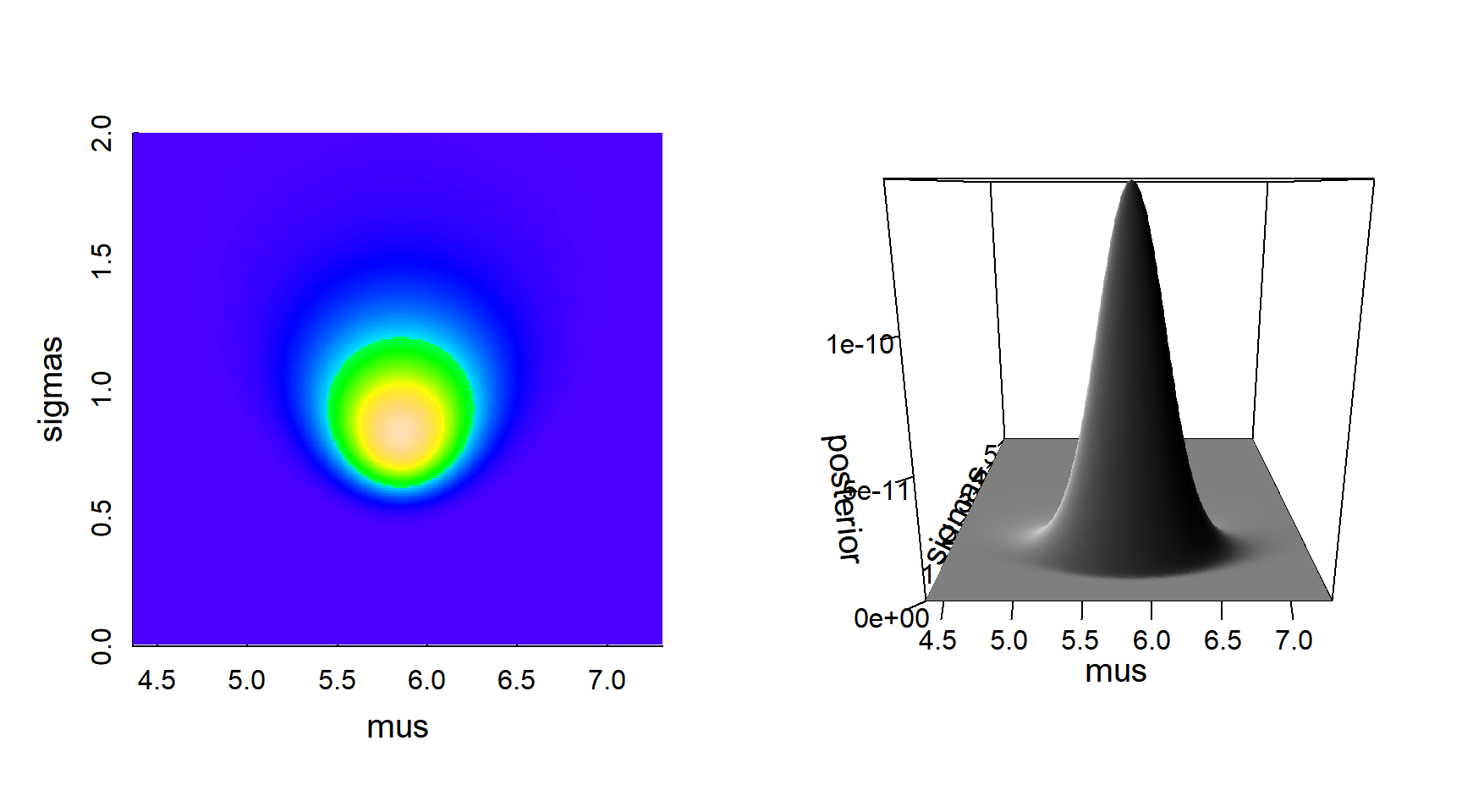
The posterior distribution seems nicely peaked.
Bayesian point estimate
Let’s find the peak (mode) of this distribution, which is one kind of central point estimate of the parameters:
(max.indices <- which(posterior == max(posterior), arr.ind=TRUE))## row col
## [1,] 190 118mu.mode <- mus[max.indices[1]]
sigma.mode <- sigmas[max.indices[2]]Lets look at the posterior distributions at the maxima, i.e. \(\pi(\mu | \sigma =\widehat\sigma, {\bf X})\) and \(\pi(\sigma | \mu = \widehat{\mu}, {\bf X}\)), and compare it to our prior distributions
mu.post <- posterior[,max.indices[2]] %>% {./sum(.)/dmu}
sigma.post <- posterior[max.indices[1],] %>% {./sum(.)/dsigma}
plot(mus, mu.post, type="l", xlim=range(X) + c(-.5,.5), lwd=2, col="darkgrey")
curve(dunif(x, mu.min, mu.max), add=TRUE, col=2, n=1e5)
legend("topleft", col=c("red", "darkgrey"), legend = c("prior", "posterior"), lty=1, bty="n", lwd=2)
plot(sigmas, sigma.post, type="l", lwd=2, col="darkgrey")
curve(dexp(x, lambda)*1e3, add=TRUE, col=2, n=1e5)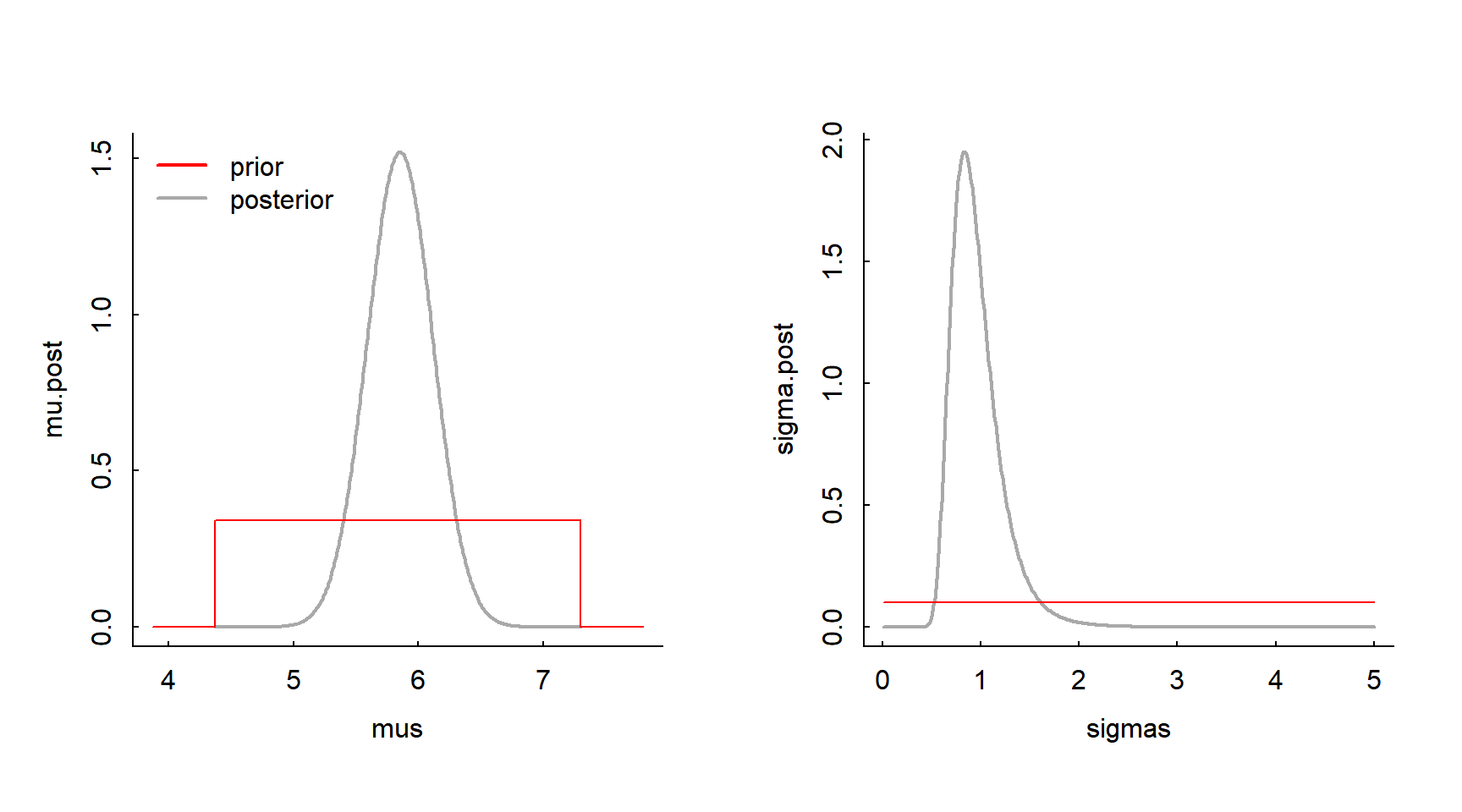
The posteriors are sharper than the priors, meaning the data have successfully “narrowed” the prior towards a useful result.
Credible intervals
A posterior distribution is the complete description of our estimates. But typically, we want to report a point estimate and (e.g.) a 95% credible interval.
To compute these, we need to integrate our posterior and find the location where the cumulative distribution is equal to 0.025 and 0.975, i.e. solve: \[\int_{\theta}^{\{CI_{low}, CI_{high}\}} \pi(\theta|{\bf X}) d\theta = \{0.025, 0.975\}\]
mu.CI.low <- mus[which.min(abs(cumsum(mu.post)*dmu - 0.025))]
mu.CI.high <- mus[which.min(abs(cumsum(mu.post)*dmu - 0.975))]
sigma.CI.low <- sigmas[which.min(abs(cumsum(sigma.post)*dsigma - 0.025))]
sigma.CI.high <- sigmas[which.min(abs(cumsum(sigma.post)*dsigma - 0.975))]Report results:
bayes.fit <- rbind(c(mu.mode, mu.CI.low, mu.CI.high), c(sigma.mode, sigma.CI.low, sigma.CI.high)) %>% data.frame
names(bayes.fit) <- c("mode", "CI.low", "CI.high")
row.names(bayes.fit) <- c("mu", "sigma")
bayes.fit## mode CI.low CI.high
## mu 6.69 5.95 7.42
## sigma 1.18 0.85 2.27Compare to MLE’s
Shall we compare this to the maximum likelihood estimators? These are (analytically solved):
\[\widehat{\mu} = \overline{X} = {1\over n} \sum_{i=1}^n X_i \] \[\widehat{\sigma} = \sqrt{{1 \over n} \sum_{i=1}^n (X_i - \overline{X})^2}\]
(mle.hat <- c(mu = mean(X), sigma = sqrt(sum((X - mean(X))^2)/length(X))))## mu sigma
## 6.689000 1.182806We can bootstrap the confidence intervals:
mle.bs <- matrix(NA, 2, 1e4)
for(i in 1:1e4){
X.resample <- sample(X, replace=TRUE)
mle.bs[, i] <- c(mu = mean(X.resample), sigma = sqrt(sum((X.resample - mean(X.resample))^2/length(X.resample))))
}
CIs <- apply(mle.bs, 1, quantile, p = c(0.025, 0.975))
(mle.fit <- cbind(mle.hat, t(CIs)))## mle.hat 2.5% 97.5%
## mu 6.689000 5.9430000 7.436050
## sigma 1.182806 0.6701629 1.485074Compare with:
bayes.fit## mode CI.low CI.high
## mu 6.69 5.95 7.42
## sigma 1.18 0.85 2.27The point estimates should be right on for both parameters, and the bootstrapped confidence intervals (maybe: BCI) and Bayesian credible intervals (maybe also: BCI?!) are right on for the means, but might be rather different for the standard deviations.
In both analyses, we can actually visualize the distribution of our estimates:
hist(mle.bs[1,], breaks=50, col="grey", bor="darkgrey", freq=FALSE, xlab="mean", main = "")
lines(mus, mu.post, lwd=2, col=2, xpd=0)
legend("topleft", legend = c("bootstrap", "posterior"),
col = c(NA, "red"), pt.bg = c("grey", NA), pch = c(22, NA), pt.cex=2, lty=c(NA, 1), bty="n")
hist(mle.bs[2,], breaks=50, col="grey", bor="darkgrey", freq=FALSE,
xlim=c(0,2.0), xlab="standard deviation", main="")
lines(sigmas, sigma.post, lwd=2, col=2)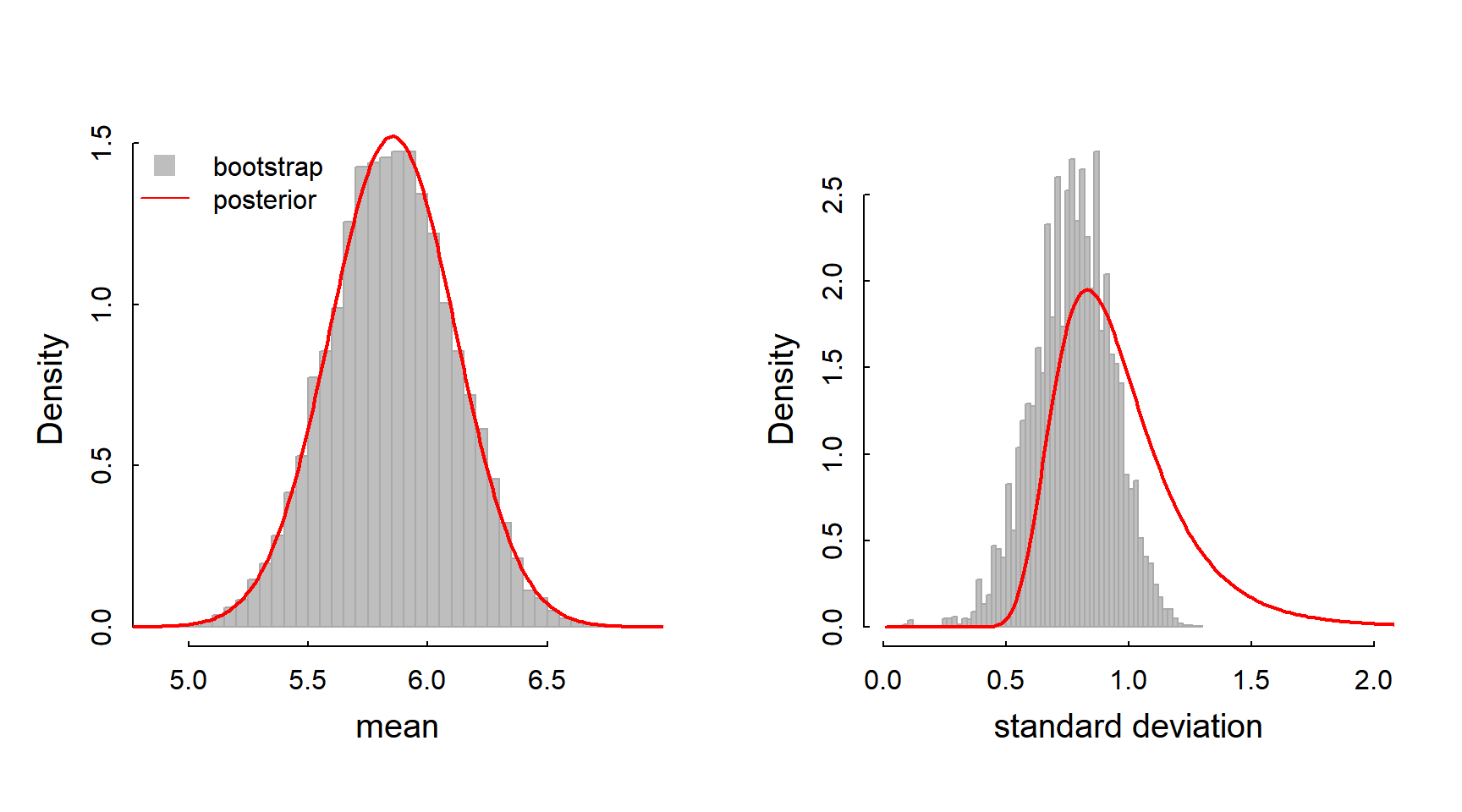
The mean distributions should look quite similar, whereas the bootstrapped distribution around \(\widehat{\sigma}\) might be multimodal, or very skewed, or generally be dissimilar to the posterior. Can you think why this might be? Which of these do you “trust” more?
Take home messages
The procedure here works and (in this simple example) provides results that are fairly consistent with the maximum likelihood estimate. Note that any reasonable prior distribution on either parameter would have worked just as well. In contrast, an analytical approach to obtaining Bayesian estimates of the mean and standard deviation is possible if you use a normal prior for \(\mu\) and a gamma prior for \(1/\sigma^2\). Since we performed the multiplication numerically, we didn’t have to worry about nice mathematical properties.
The disadvantage of this approach is that it requires a fairly fine “mesh” for the parameters, and is usually prohibitively slow if there are many more parameters. Note that the actual multiplication (“updating” in Bayesian jargon) step that we used here (in the outer() command) takes a noticable amount of time. Make that array a four or five-dimensional array, and the order of computation becomes infeasible … and many real, relevant models have many more parameters than that.
The - rather clever - solution to this problem is the topic of the final lecture and lab.