Biol709 / Bsci339 Final Thoughts
Elie Gurarie
January 22, 2018
What have we learned?
- Basic data wrangling
- incl. regression coefs / sums of squares
- Probability theory
- distributions!
- Foundations of Inference
- hypothesis tests / confidence intervals
- Linear modeling / ANOVA
- (relies on pieces of everthing above!)
- Likelihoods and MLE’s
- … under the hood of GLM and lots more
- Generalizing linear models
- GLS (dependent data)
- random coefficients (LME’s)
- Bayesian Inference
- theory
- MCMC (with STAN)
Some Books



What we didn’t learn: More Advanced Regression
- GLMM
- easy!
- Additive models (GAM’s)
- leading to Generalized Linear Additive Mixed Models (GLAMM)
- Phylogenetic correlations
- easy! … just add the relationship matrix to `gls``
- Lasso / Spline bases / Ridge-regression
- fancy - but effective - stuff for solving some practical problems with outliers / funny distributions / non-linear responses.

What we didn’t learn: Multivariate Stats
For analyzing data with complex (multivariate) responses - e.g. multiple species presence / absence data.
- Multivariate regression
- MANOVA / MANCOVA
- Canonical-correlation analysis (CCA)
- Ordination / Dimension Reduction
- Principle Components (PCA)
- Factor Analysis
- Non-metric multidimensional scaling

What we didn’t learn: Machine Learning / Prediction
Broadly : Analysis that minimizes distributional assumptions and does NOT have inference as a goal, but only Prediction
- Nearest neighbor
- Random forests
- Clustering
- Cross-validation
- Dimension reduction
Actually: LOTS of overlap with topics we’ve discussed
(more than often acknowledged).

===========================================
My philosophy (usually):
- Biologically meaningful parameters
- As mechanistic (as possible) models that allow for random processes through a probability model
- Before using a new tool … have some idea what’s going on beneath the hood
My goal in this class:
To empower you to not only peek under hoods, but create your own crazy original analysis machines!
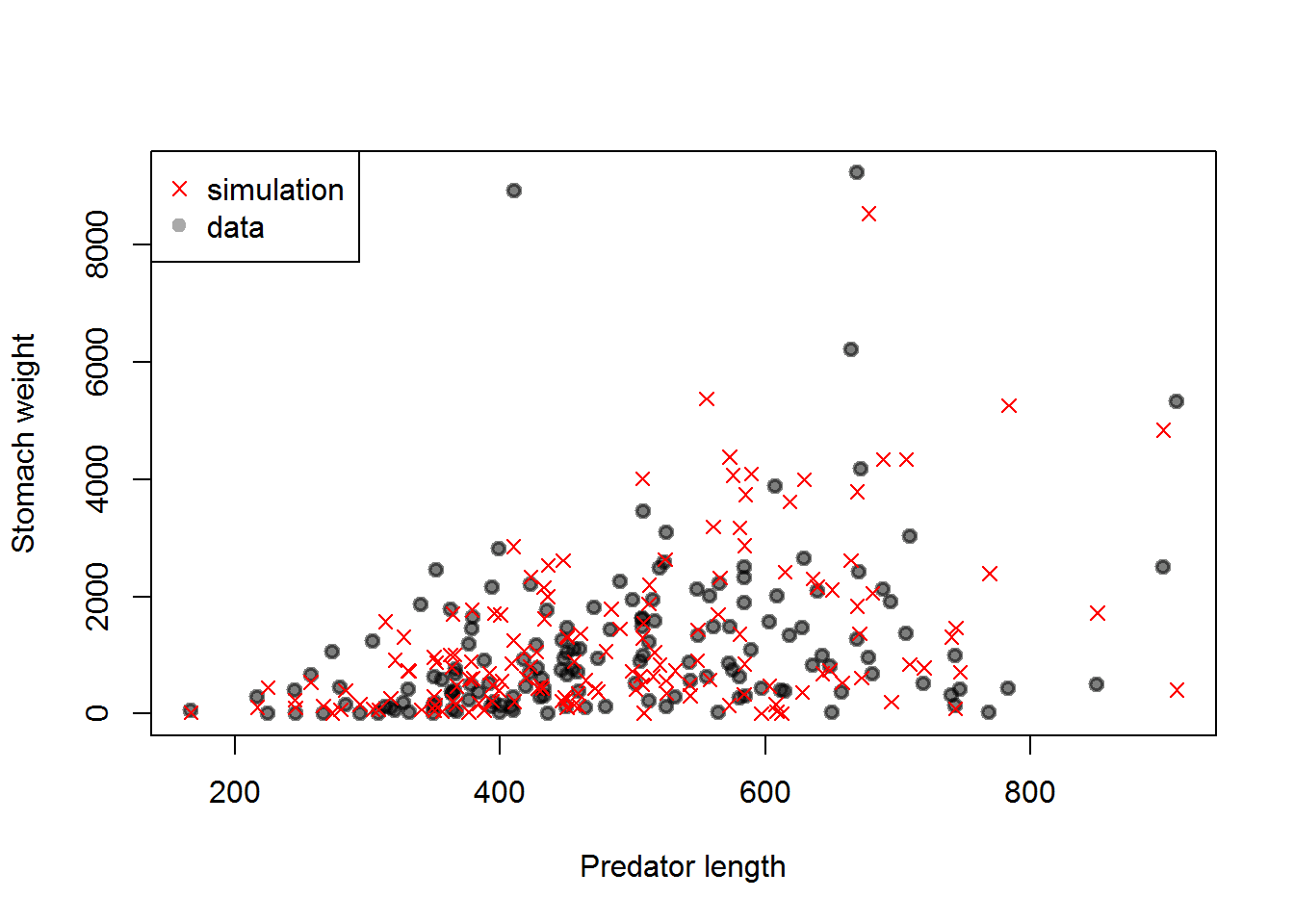
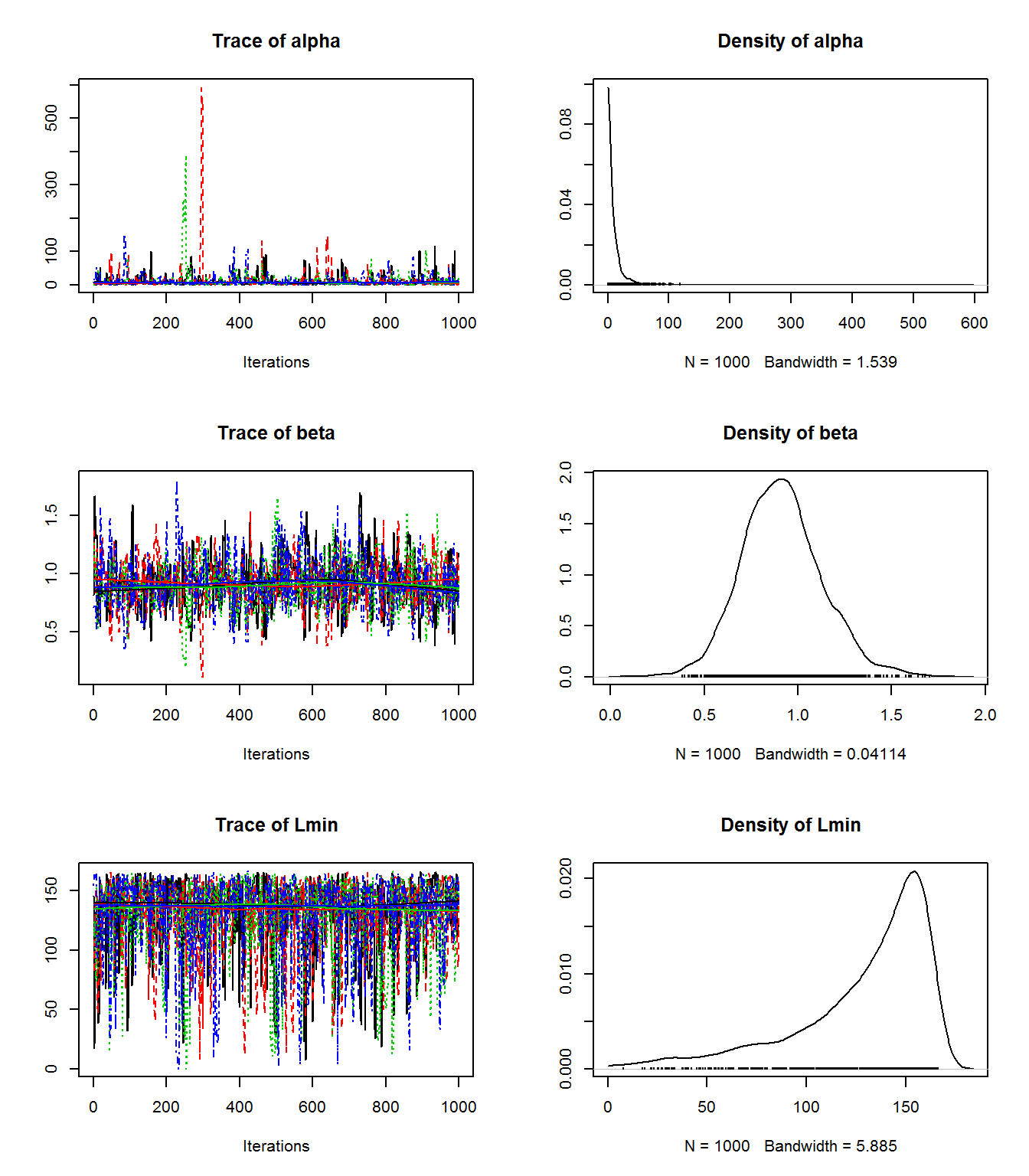
Atlantic Cod Stomach Example
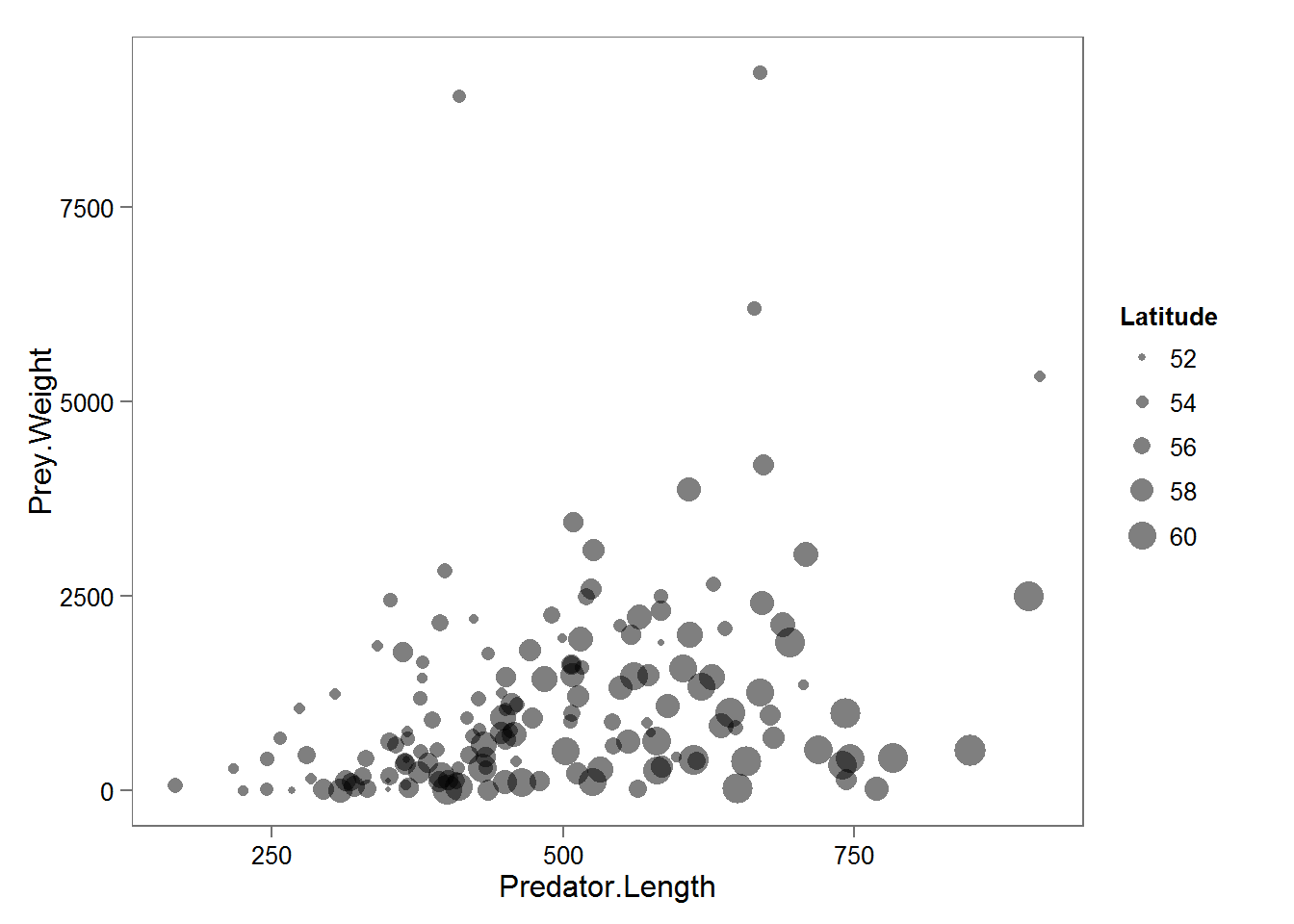
Model
\[W_{mean} = (\alpha_0 + \alpha_1 \text{Latitude})^\beta \times (L - L_{min})\] \[W_{obs} \sim \text{Exp}(mean = W_{mean})\]
where
- \(W\) is average stomach size
- \(\alpha\)’s are intercept / slope of regression against Latitude
- \(\beta\) is a scaling paramter
- \(L_min\) is the minimum size of a (captured) cod

Altantic Cod Stan Code
data {
int n; //
real Length[n]; // Predator length
real WeightObs[n]; // Observed weight of stomach contents
real Latitude[n]; // Latitudes at observations
}
parameters{
real alpha0; // intercept & ...
real alpha1; // ... slope of latitude dependence
real Lmin; // minimum length of predator for stomach contents
}
model{
real WeightMean[n]; // mean weight
// Priors
alpha0 ~ normal(0,1e3);
alpha1 ~ normal(0,1e3);
Lmin ~ uniform(0, min(Length));
// Likelihood
for(i in 1:n){
WeightMean[i] <- (alpha0 + alpha1*Latitude[i]) * (Length[i]-Lmin);
WeightObs[i] ~ exponential(1/WeightMean[i]);
}
}Atlantic Cod Stomach Example
With that, Welcome to ….
The SECOND Annual Mega-Mini-Symposium on Data Analysis and Modeling in Ecology and Environmental Sciences and Genetics And Random Stuff
- Opening Remarks and Welcome
- Talks
First Speaker on First Topic
Second Speaker on Second Topic
Third Speaker on Third Topic
Fourth Speaker on Fourth Topic
etc…

Rubric
For each presentation, please write a brief sentence or two answering the following questions:

5. Comment on at least ONE result